-
PDF
- Split View
-
Views
-
Cite
Cite
Ishrat Maliha Islam, Ted Erclik, Imp and Syp mediated temporal patterning of neural stem cells in the developing Drosophila CNS, Genetics, Volume 222, Issue 1, September 2022, iyac103, https://doi.org/10.1093/genetics/iyac103
Close - Share Icon Share
Abstract
The assembly of complex neural circuits requires that stem cells generate diverse types of neurons in the correct temporal order. Pioneering work in the Drosophila embryonic ventral nerve cord has shown that neural stem cells are temporally patterned by the sequential expression of rapidly changing transcription factors to generate diversity in their progeny. In recent years, a second temporal patterning mechanism, driven by the opposing gradients of the Imp and Syp RNA-binding proteins, has emerged as a powerful way to generate neural diversity. This long-range temporal patterning mechanism is utilized in the extended neural stem cell lineages of the postembryonic fly brain. Here, we review the role played by Imp and Syp gradients in several neural stem cell lineages, focusing on how they specify sequential neural fates through the post-transcriptional regulation of target genes, including the Chinmo and Mamo transcription factors. We further discuss how upstream inputs, including hormonal signals, modify the output of these gradients to couple neurogenesis with the development of the organism. Finally, we review the roles that the Imp and Syp gradients play beyond the generation of diversity, including the regulation of stem cell proliferation, the timing of neural stem cell lineage termination, and the coupling of neuronal birth order to circuit assembly.
Introduction
The assembly of complex neuronal circuits requires that neural stem cells (NSCs) generate diverse types of neurons and coordinately regulate their number and location. Over the last 30 years, the Drosophila central nervous system (CNS) has emerged as a powerful model in which to study the patterning mechanisms that regulate neurogenesis. In the developing fly, NSCs, termed neuroblasts (NBs), integrate spatial and temporal patterning inputs to generate neurons with unique positional, stoichiometric, morphological, and functional characteristics. The spatial and temporal patterning of NBs have been best studied in the embryonic ventral nerve cord where, in the spatial axis, segment polarity, Hox, and dorsal-ventral genes combine to give each NB a unique positional identity (Skeath et al., 1995; McDonald et al., 1998; Technau et al., 2006; Karlsson et al., 2010). In the temporal axis, NBs sequentially express the temporal transcription factors (tTFs) Hunchback, Kruppel, Pdm1/2, Castor, and Grainyhead to generate birth-order-dependent neuronal diversity (Isshiki et al., 2001). Extensive cross-regulation between the tTFs drives the intrinsic clock forward, such that the majority of these temporal windows last for only 1–2 cell divisions (Isshiki et al., 2001; Doe, 2017; Miyares and Lee, 2019). These rapid transitions between tTF windows allow for the generation of a large amount of neuronal diversity over a short time period. The independently acting spatial and temporal patterning inputs are integrated by the NB, at both the epigenetic and transcriptional levels, to regulate multiple aspects of neurogenesis, including neuronal identity and stem cell proliferation mode (Baumgardt et al., 2014; Sen et al., 2019; Ray et al., 2022). Integration of spatial and temporal patterning has since been shown in the larval optic lobe (Li et al., 2013; Bertet et al., 2014; Erclik et al., 2017; Konstantinides et al., 2021; Zhu et al., 2022), where the observation that optic lobe NBs use an entirely different set of tTFs suggests that the temporal patterning of NSCs is a general mechanism used to generate diversity. Indeed, in the developing vertebrate nervous system, where spatial patterning has long had an established role in neurogenesis, temporal patterning has emerged as critical for the generation of diversity in the retina, spinal cord, and cerebral cortex (Dessaud et al., 2008; Naka et al., 2008; Trimarchi et al., 2008; Mattar et al., 2015; Clark et al., 2019; Delile et al., 2019; Telley et al., 2019).
In recent years, a second temporal patterning mechanism has emerged as a powerful way to generate neuronal diversity. This mode of temporal patterning, which functions in the extended postembryonic NB lineages of the larval and pupal CNS, utilizes slowly progressing gradients of proteins to generate diversity. Unlike the short-range rapid transitions driven by tTFs in the embryonic NBs, the protracted nature of this longer-range patterning mechanism allows for the generation of greater numbers of neurons with identical fates to meet the needs of large and complex adult circuits. Moreover, the extended duration of patterning over days allows for the integration of extrinsic cues to couple neurogenesis with organismal development. Here, we review how opposing gradients of the IGF-II mRNA-binding protein (Imp) and Syncrip (Syp) RNA-binding proteins, and their downstream transcription factors, pattern postembryonic NB lineages to generate neuronal diversity. We discuss how cell-extrinsic signals, including ecdysone and Activin signaling, modify the timing and output of these gradients and explore the intriguing possibility that the tTF- and gradient-based temporal mechanisms may act concurrently to increase diversity. Finally, we review roles for these gradients beyond diversity, including in the regulation of NB growth and termination and in the coordinated assembly of neural circuits.
Opposing Imp and Syp gradients temporally pattern postembryonic NB lineages
The gradient-based temporal patterning of NBs has been described in several lineages of the postembryonic central brain and ventral nerve cord. The majority of NBs in these lineages divide up to 50 times over the course of 4 days during larval and pupal development (Truman and Bate, 1988; Doe, 2017). The increased number of these stem cell divisions necessitates a patterning mechanism that works over a longer timespan than the short-range tTF cascades described in the embryo. Here, we describe how gradients of the Imp and Syp RNA-binding proteins generate neural diversity in the 2 postembryonic lineages where they have been best studied, the mushroom body and Type II NBs of the central brain.
Imp and Syp patterning of mushroom body NBs
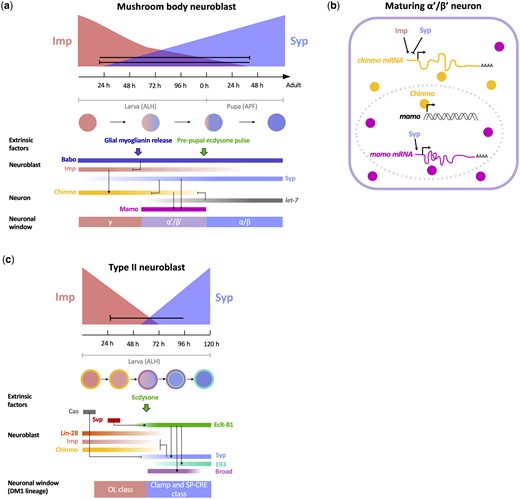
The neurons of the mushroom body, the primary center for learning and memory in the fly brain, are generated by 4 NBs that divide continuously throughout larval and pupal development (Lee et al., 1999). These NBs generate over 2000 neurons belonging to 3 major types; γ neurons made between the first and mid-third larval instars, α’/β’ neurons made from the mid-third larval instar to the beginning of pupation, and α/β neurons made from pupation until eclosion. In a search for genes that regulate the sequential production of these neurons, transcriptomic analysis of mushroom body NBs at several developmental stages identified 2 RNA-binding proteins, Imp and Syp, that are expressed in complimentary gradients (Liu et al., 2015; Fig. 1a). Imp is highly expressed in early NBs where it is required for the specification of γ neurons, whereas the peak of Syp expression occurs in late NBs, where it is required for α/β fates. In intermediate-aged NBs, in which the genes are coexpressed, Imp and Syp are both required for α’/β’ neuronal specification. Functional analyses have shown that the Imp and Syp gradient progressions are in part driven by cross-repression between the 2 genes; the removal of Syp leads to the extended expression of Imp, and the loss of Imp leads to the precocious expression of Syp (Liu et al., 2015).
Temporal gradient patterning of postembryonic NBs. a) Imp and Syp are expressed in opposing gradients and repress each other in mushroom body NBs. In nascent neurons, Imp promotes Chinmo expression, while Syp represses it, which results in a declining gradient of Chinmo levels in mushroom body neurons; γ neurons express high levels of Chinmo, α’/β’ neurons express low levels of Chinmo, and α/β neurons do not express Chinmo. Low Chinmo and Syp promote the expression of Mamo, which specifies the α’/β’ fate. Extrinsic activin and ecdysone signaling regulate the timing of neuronal temporal transitions. Activin promotes the switch from γ to α’/β’ fates by downregulating Imp expression through the Babo receptor. Ecdysone signaling in the prepupa regulates the switch between α’/β’ and α/β fates via activation of the let-7 miRNA, which represses chinmo. b) Regulation of α’/β’ neuronal fate specification. In a newborn α’/β’ neuron, Imp promotes, while Syp represses, chinmo mRNA translation, likely through direct binding of its 5′ UTR. The resulting low levels of Chinmo expression allow for the activation of mamo transcription. Syp additionally stabilizes mamo mRNA transcripts to promote Mamo expression. c) Opposing Imp and Syp temporal gradients pattern Type II central brain NBs. Imp, Chinmo and Lin-28 are expressed in NBs early, while Syp, E93, and Broad are expressed late. Cas is expressed very early and represses Syp when its expression is prolonged. Svp upregulates EcR-B1, which results in the switch from early to late gene expression at 60 h ALH. Syp is required for the repression of Imp and Chinmo but is not required for Broad and E93 expression. Temporally regulated genes in the NB generate early and late neuronal specification windows. In the DM1 lineage, Imp-dependent OL class neurons are born early and Syp-dependent Clamp/SP-CRE class neurons are born late.
How do RNA-binding proteins regulate cell fate specification? Imp and Syp are inherited in mushroom body neurons where they are required for the temporally restricted expression of 2 BTB zinc-finger transcription factors, Chronologically inappropriate morphogenesis (Chinmo) and Maternal gene required for meiosis (Mamo; Zhu et al., 2006; Liu et al., 2019, 2015; Fig. 1a). Chinmo is expressed in mushroom body neurons in a declining temporal gradient that parallels that of Imp (Zhu et al., 2006). High levels of Chinmo in early-born neurons specify the γ fate, whereas lower levels in intermediately generated neurons specify the subsequent α’/β’ fate. In late-born neurons, the absence of Chinmo expression is required for the specification of the α/β fate. chinmo transcripts are present at comparable levels in neurons born during all 3 developmental stages, which suggests that post-transcriptional regulation of chinmo mRNA establishes the Chinmo protein gradient. Indeed, Imp and Syp post-transcriptionally regulate chinmo transcripts, likely via binding at the 5′ UTR, with Imp promoting Chinmo translation and Syp repressing it (Zhu et al., 2006; Liu et al., 2015). The resulting early/high Chinmo vs intermediate/low Chinmo windows lead to the specification of 2 distinct cell types via the differential activation of Mamo (Liu et al., 2019; Fig. 1b). Only the low levels of Chinmo that are present during the intermediate window are able to activate the transcription of mamo, which subsequently acts as a terminal selector gene to specify the α’/β’ fate. Syp further consolidates the α’/β’ fate by promoting the maturation and/or stabilization of mamo transcripts. The Imp- and Syp-mediated post-transcriptional regulation of chinmo and mamo illustrates how RNA-binding proteins can act at multiple levels to specify temporally restricted neuronal fates. Moreover, the dual role of Syp as an indirect activator of mamo (through Chinmo) and direct stabilizer of mamo transcripts ensures that the temporal patterning of the α’/β’ fate is a robust process with multiple safeguards.
Future analyses should focus on the mechanism by which Imp and Syp regulate Chinmo and Mamo expression. These studies could include RNA-binding assays, such as RNA immunoprecipitation and sequencing (RIPseq), to confirm that Imp and Syp directly bind to chinmo and mamo transcripts. RIPseq may also allow for the identification of additional factors that are post-transcriptionally regulated by Imp and Syp to specify neuronal fates (Table 1). For example, are there transcription factors that act as terminal selectors for the γ and α/β fates? It will also be interesting to determine if additional temporal windows are present in the mushroom body lineage. Recent morphological and transcriptomic studies have shown that the γ and α’/β’ neurons can each be divided into 2 subtypes, while α/β neurons are comprised of 3 distinct types of neurons (Shih et al., 2019). Indeed, one subtype of α/β neuron, the α/β pioneer, is born in a short temporal window located between the α’/β’ and α/β windows (Lee et al., 1999; Zhu et al., 2003). Are these subtypes each dependent on unique levels of Imp and Syp for their specification? Identifying unique markers for each of the potentially 7 neuronal subtypes will facilitate these analyses. One transcription factor that may diversify the α/β class is E93 (Pahl et al., 2019), which is expressed in neurons born in the second half of the α/β window.
Imp and Syp targets in Drosophila NB lineages.
| Gene . | Temporal patterning function . | Mode of Imp/Syp regulation . |
|---|---|---|
| Chronologically inappropriate morphogenesis (Chinmo) | Transcription factor. Expressed in a descending temporal gradient in NBs and neurons to promote early and intermediate fates. Low levels activate Mamo to generate α’/β’ neurons in the MB. | Imp promotes Chinmo mRNA translation in MB neurons. Syp represses Chinmo post-transcriptionally. Regulation in MB likely via 5′ UTR. Bound by Imp and Syp in affinity pull-down assay.a Direct target of Imp in larval brain RIPseq.b |
| Maternal gene required for meiosis (Mamo) | Transcription factor. Required in MB neurons for α’/β’ fate specification. | Syp stabilizes Mamo mRNA to promote protein expression in α’/β’ neurons. Direct target of Imp in larval brain RIPseq.b |
| Myc (Myc) | Transcription factor. Promotes growth and proliferation in young NBs. | Imp stabilizes Myc mRNA to promote expression in the NB. Direct target of Imp in larval brain RIPseq.b |
| Prospero (Pros) | Transcription factor. Promotes cell cycle exit in late-stage Type I and II NBs. | Syp stabilizes Pros mRNA to promote accumulation of Pros protein in the NB nucleus. Bound by Syp in Co-IP of larval brain lysates.c Direct target of Imp in larval brain RIPseq.b |
| Ecdysone-induced protein 93F (E93) | Transcription factor. Promotes NB decommissioning in the MB via PI3-kinase-mediated autophagy. Expression marks a late temporal window in Type II NBs. | Imp represses E93 expression in early MB NBs. Syp promotes E93 expression in late MB NBs. Direct target of Imp in larval brain RIPseq.b |
| Mediator complex components (Med6, Med27, and Med31) | Subunits of the mediator complex. Act with Ecdysone signaling to change energy metabolism and slow growth in late-stage NBs. | Imp binds to Med6 mRNA and negatively regulates its translation. Bound by Imp in Co-IP of larval brain lysates.d |
| IGF-II mRNA-binding protein (Imp) | RNA-binding protein. Expressed in a descending temporal gradient in NBs and neurons to promote early and intermediate fates. Promotes NB growth and proliferation in early larval NBs. | In MB NBs, Imp and Syp reciprocally repress each other. In non-MB NBs, Syp represses Imp. Targets itself in larval brain RIPseq.b |
| Syncrip (Syp) | RNA-binding protein. Expressed in an ascending temporal gradient in NBs and neurons to promote intermediate and late fates. Promotes NB decommissioning and lineage termination. | In MB NBs, Imp and Syp reciprocally repress each other. In non-MB NBs, Imp does not repress Syp. Direct target of Imp in larval brain RIPseq.b |
| Gene . | Temporal patterning function . | Mode of Imp/Syp regulation . |
|---|---|---|
| Chronologically inappropriate morphogenesis (Chinmo) | Transcription factor. Expressed in a descending temporal gradient in NBs and neurons to promote early and intermediate fates. Low levels activate Mamo to generate α’/β’ neurons in the MB. | Imp promotes Chinmo mRNA translation in MB neurons. Syp represses Chinmo post-transcriptionally. Regulation in MB likely via 5′ UTR. Bound by Imp and Syp in affinity pull-down assay.a Direct target of Imp in larval brain RIPseq.b |
| Maternal gene required for meiosis (Mamo) | Transcription factor. Required in MB neurons for α’/β’ fate specification. | Syp stabilizes Mamo mRNA to promote protein expression in α’/β’ neurons. Direct target of Imp in larval brain RIPseq.b |
| Myc (Myc) | Transcription factor. Promotes growth and proliferation in young NBs. | Imp stabilizes Myc mRNA to promote expression in the NB. Direct target of Imp in larval brain RIPseq.b |
| Prospero (Pros) | Transcription factor. Promotes cell cycle exit in late-stage Type I and II NBs. | Syp stabilizes Pros mRNA to promote accumulation of Pros protein in the NB nucleus. Bound by Syp in Co-IP of larval brain lysates.c Direct target of Imp in larval brain RIPseq.b |
| Ecdysone-induced protein 93F (E93) | Transcription factor. Promotes NB decommissioning in the MB via PI3-kinase-mediated autophagy. Expression marks a late temporal window in Type II NBs. | Imp represses E93 expression in early MB NBs. Syp promotes E93 expression in late MB NBs. Direct target of Imp in larval brain RIPseq.b |
| Mediator complex components (Med6, Med27, and Med31) | Subunits of the mediator complex. Act with Ecdysone signaling to change energy metabolism and slow growth in late-stage NBs. | Imp binds to Med6 mRNA and negatively regulates its translation. Bound by Imp in Co-IP of larval brain lysates.d |
| IGF-II mRNA-binding protein (Imp) | RNA-binding protein. Expressed in a descending temporal gradient in NBs and neurons to promote early and intermediate fates. Promotes NB growth and proliferation in early larval NBs. | In MB NBs, Imp and Syp reciprocally repress each other. In non-MB NBs, Syp represses Imp. Targets itself in larval brain RIPseq.b |
| Syncrip (Syp) | RNA-binding protein. Expressed in an ascending temporal gradient in NBs and neurons to promote intermediate and late fates. Promotes NB decommissioning and lineage termination. | In MB NBs, Imp and Syp reciprocally repress each other. In non-MB NBs, Imp does not repress Syp. Direct target of Imp in larval brain RIPseq.b |
Imp and Syp targets in Drosophila NB lineages.
| Gene . | Temporal patterning function . | Mode of Imp/Syp regulation . |
|---|---|---|
| Chronologically inappropriate morphogenesis (Chinmo) | Transcription factor. Expressed in a descending temporal gradient in NBs and neurons to promote early and intermediate fates. Low levels activate Mamo to generate α’/β’ neurons in the MB. | Imp promotes Chinmo mRNA translation in MB neurons. Syp represses Chinmo post-transcriptionally. Regulation in MB likely via 5′ UTR. Bound by Imp and Syp in affinity pull-down assay.a Direct target of Imp in larval brain RIPseq.b |
| Maternal gene required for meiosis (Mamo) | Transcription factor. Required in MB neurons for α’/β’ fate specification. | Syp stabilizes Mamo mRNA to promote protein expression in α’/β’ neurons. Direct target of Imp in larval brain RIPseq.b |
| Myc (Myc) | Transcription factor. Promotes growth and proliferation in young NBs. | Imp stabilizes Myc mRNA to promote expression in the NB. Direct target of Imp in larval brain RIPseq.b |
| Prospero (Pros) | Transcription factor. Promotes cell cycle exit in late-stage Type I and II NBs. | Syp stabilizes Pros mRNA to promote accumulation of Pros protein in the NB nucleus. Bound by Syp in Co-IP of larval brain lysates.c Direct target of Imp in larval brain RIPseq.b |
| Ecdysone-induced protein 93F (E93) | Transcription factor. Promotes NB decommissioning in the MB via PI3-kinase-mediated autophagy. Expression marks a late temporal window in Type II NBs. | Imp represses E93 expression in early MB NBs. Syp promotes E93 expression in late MB NBs. Direct target of Imp in larval brain RIPseq.b |
| Mediator complex components (Med6, Med27, and Med31) | Subunits of the mediator complex. Act with Ecdysone signaling to change energy metabolism and slow growth in late-stage NBs. | Imp binds to Med6 mRNA and negatively regulates its translation. Bound by Imp in Co-IP of larval brain lysates.d |
| IGF-II mRNA-binding protein (Imp) | RNA-binding protein. Expressed in a descending temporal gradient in NBs and neurons to promote early and intermediate fates. Promotes NB growth and proliferation in early larval NBs. | In MB NBs, Imp and Syp reciprocally repress each other. In non-MB NBs, Syp represses Imp. Targets itself in larval brain RIPseq.b |
| Syncrip (Syp) | RNA-binding protein. Expressed in an ascending temporal gradient in NBs and neurons to promote intermediate and late fates. Promotes NB decommissioning and lineage termination. | In MB NBs, Imp and Syp reciprocally repress each other. In non-MB NBs, Imp does not repress Syp. Direct target of Imp in larval brain RIPseq.b |
| Gene . | Temporal patterning function . | Mode of Imp/Syp regulation . |
|---|---|---|
| Chronologically inappropriate morphogenesis (Chinmo) | Transcription factor. Expressed in a descending temporal gradient in NBs and neurons to promote early and intermediate fates. Low levels activate Mamo to generate α’/β’ neurons in the MB. | Imp promotes Chinmo mRNA translation in MB neurons. Syp represses Chinmo post-transcriptionally. Regulation in MB likely via 5′ UTR. Bound by Imp and Syp in affinity pull-down assay.a Direct target of Imp in larval brain RIPseq.b |
| Maternal gene required for meiosis (Mamo) | Transcription factor. Required in MB neurons for α’/β’ fate specification. | Syp stabilizes Mamo mRNA to promote protein expression in α’/β’ neurons. Direct target of Imp in larval brain RIPseq.b |
| Myc (Myc) | Transcription factor. Promotes growth and proliferation in young NBs. | Imp stabilizes Myc mRNA to promote expression in the NB. Direct target of Imp in larval brain RIPseq.b |
| Prospero (Pros) | Transcription factor. Promotes cell cycle exit in late-stage Type I and II NBs. | Syp stabilizes Pros mRNA to promote accumulation of Pros protein in the NB nucleus. Bound by Syp in Co-IP of larval brain lysates.c Direct target of Imp in larval brain RIPseq.b |
| Ecdysone-induced protein 93F (E93) | Transcription factor. Promotes NB decommissioning in the MB via PI3-kinase-mediated autophagy. Expression marks a late temporal window in Type II NBs. | Imp represses E93 expression in early MB NBs. Syp promotes E93 expression in late MB NBs. Direct target of Imp in larval brain RIPseq.b |
| Mediator complex components (Med6, Med27, and Med31) | Subunits of the mediator complex. Act with Ecdysone signaling to change energy metabolism and slow growth in late-stage NBs. | Imp binds to Med6 mRNA and negatively regulates its translation. Bound by Imp in Co-IP of larval brain lysates.d |
| IGF-II mRNA-binding protein (Imp) | RNA-binding protein. Expressed in a descending temporal gradient in NBs and neurons to promote early and intermediate fates. Promotes NB growth and proliferation in early larval NBs. | In MB NBs, Imp and Syp reciprocally repress each other. In non-MB NBs, Syp represses Imp. Targets itself in larval brain RIPseq.b |
| Syncrip (Syp) | RNA-binding protein. Expressed in an ascending temporal gradient in NBs and neurons to promote intermediate and late fates. Promotes NB decommissioning and lineage termination. | In MB NBs, Imp and Syp reciprocally repress each other. In non-MB NBs, Imp does not repress Syp. Direct target of Imp in larval brain RIPseq.b |
Imp and Syp patterning of Type II NBs
Opposing Imp and Syp temporal gradients have subsequently been shown to pattern all postembryonic central brain and ventral nerve cord lineages in which they have been analyzed (Ren et al., 2017; Syed et al., 2017). Their role outside of the mushroom body has been best studied in the 16 Type II NBs of the central brain, which generate neurons that innervate several regions of the CNS, including the fan-shaped body, optic lobe, and protocerebrum. Drosophila NBs can be characterized by their mode of division: Type I NBs, which comprise the majority of NBs (including those of the mushroom body), asymmetrically divide to self-renew, and generate a ganglion mother cell (GMC), which subsequently divides to generate 2 postmitotic neurons or glia; in contrast, Type II NBs asymmetrically divide to generate an intermediate neural progenitor (INP), which then undergoes multiple divisions to produce a short lineage comprised of ∼6 GMCs (Bello et al., 2008; Bayraktar and Doe, 2013; Viktorin et al., 2013). In Type II lineages, Imp and Syp regulate fates in a manner similar to their role in the mushroom body; Imp promotes early fates and Syp promotes late ones (Ren et al., 2017; Syed et al., 2017; Fig. 1c). For example, in the DM1 Type II NB lineage, knocking down Imp results in the loss of early-born optic lobe innervating neurons, and the expansion of late-born Clamp and SP-CRE neurons (Ren et al., 2017). Syp loss-of-function results in a reciprocal change, with the early optic lobe fates expanded at the expense of the late-born Clamp neurons. Intriguingly, middle-born fates are expanded in Imp or Syp overexpression mutants. This observation suggests that the coexpression of Imp and Syp defines an intermediate temporal window, which may be analogous to the specification of α’/β’ neurons when Imp and Syp are coexpressed in the intermediately aged NBs of the mushroom body.
The restricted temporal expression of 6 additional DNA- and RNA-binding factors suggests that there may be up to 7 distinct temporal windows in Type II NBs (Syed et al., 2017; Fig. 1c). These temporally expressed factors can be categorized as early (coexpressed with Imp) or late (coexpressed with Syp), with the transition between the 2 occurring at ∼60 h after larval hatching (ALH). In the early window, descending gradients of Imp, Chinmo, and Lin-28 overlap with early subwindows of Castor and Seven-up (Svp) expression. In the late window, the overlapping expression of Syp, Broad, and E93 defines up to 3 potential subwindows (Broad, Broad+E93, and E93). Consistent with its role in the mushroom body, Chinmo is required for the generation of early Type II-derived neuronal fates; chinmo mutant NBs in the DM1 lineage generate a reduced number of the early-born optic lobe innervating neurons (Ren et al., 2017). However, the loss of early fates is less severe in chinmo mutants compared to those observed in the Imp loss of function, which suggests that Imp may promote early fates through additional downstream targets. Future analyses should determine whether Broad and E93 are required for the specification of late fates in the DM1 lineage. In addition, it will be interesting to determine whether Mamo is also expressed in Type II lineages, particularly in the intermediate neural progeny that coexpress Imp and Syp.
The unique mode of Type II NB divisions raises the possibility that multiple temporal patterning mechanisms act concurrently to diversify neural fates in these lineages. The INPs of Type II NBs sequentially express the tTFs Dichaete, Grainy-head, and Eyeless as they age (Bayraktar and Doe, 2013). These tTFs are inherited by the neurons to generate diversity in the progeny of an individual INP. Could the gradient patterning of Type II NBs act combinatorially with the tTF patterning of their daughter INPs to increase diversity? Intriguingly, while Imp is required for early fates and Syp is required for late ones, there is still diversity present in the neurons that are generated in these mutant backgrounds. For example, in mutant DM1 Type II NBs that are trapped in a high Imp window, multiple types of early-born optic lobe innervating neurons are still generated (Ren et al., 2017). These data are consistent with the possibility that the temporal gradient patterning in the NB assigns coarse fates to the neurons generated in each window (e.g. early optic lobe vs late central complex innervating neurons), whereas the tTF cascade that patterns the INP lineages diversifies these fates to generate neuronal subtypes (e.g. different types of early optic lobe innervating neurons). It will be interesting to determine whether the 2 temporal patterning cascades work together or independently to generate diverse fates. One possibility is that the gradient patterning of the NB generates epigenetic changes in the chromatin landscape of its INP progeny that alter the binding sites available to the tTFs. These future analyses will require the mapping of molecular markers to specific Type II-derived neuronal cell types to allow for the determination of which cell types are affected in temporal gradient and tTF mutant backgrounds. The development of additional intersectional genetic approaches to label neurons that are derived from distinct combinations of temporal windows will also facilitate future studies.
A universal role for Imp and Syp in the patterning of postembryonic NB lineages
Imp and Syp temporal patterning has been studied in several additional postembryonic lineages, including in the antero-dorsal (AD) NBs of the antennal lobe and the NBs of the ventral nerve cord (Liu et al., 2015; Doe, 2017; Allen et al., 2020). In each of the lineages examined, Imp and Chinmo gradients promote early neuronal fates, whereas subsequent Syp expression is required for late fates. This observation suggests that the Imp-Chinmo-Syp axis represents a general patterning mechanism for the temporal specification of postembryonic NB lineages. While this mechanism is similar across NB lineages, there are several key differences. The slopes of the Imp and Syp gradients vary dramatically between lineages (Fig. 1, a and c). Imp and Syp are expressed in steep, rapidly progressing gradients in Type II and AD NBs, but in shallow, slowly changing gradients in mushroom body NBs (Liu et al., 2015; Ren et al., 2017; Syed et al., 2017). The regulatory relationships between Imp, Chinmo, and Syp also differ between lineages. Imp promotes Chinmo and represses Syp expression in the mushroom body lineage, but is dispensable for Chinmo activation and Syp repression in Type II NBs (Liu et al., 2015; Syed et al., 2017). Moreover, Chinmo and Broad are expressed in the NB in Type II lineages, but are restricted to the neurons of the mushroom body lineage (Zhu et al., 2006; Maurange et al., 2008; Zhou et al., 2009; Syed et al., 2017). The effects of Imp and Syp on fate specification can also differ depending on the lineage. As described above, Imp and Syp are required for the specification of early and late fates, respectively, in mushroom body and Type II lineages (Liu et al., 2015; Ren et al., 2017; Syed et al., 2017). In the AD lineage, the knockdown of Imp or Syp alters the ratio of early vs late fates (Liu et al., 2015). However, 21 out of the 22 neuronal subtypes are still generated, which suggests that while Imp and Syp are dispensable for the generation of neural diversity, they may regulate the relative number of neurons born within each temporal window by controlling the pace of specification.
In the future, it will be important to determine how the lineage-specific differences outlined above are regulated by NB-intrinsic factors, including the transcription factors that spatially pattern NBs in the central brain. A first step may require the identification of the upstream genes that directly activate Imp, Syp, and Chinmo expression. Future analyses should also investigate the mechanisms by which the temporal transitions are regulated. Intriguingly, progression through the G1/S cell cycle checkpoint is required for the Imp–Syp transition, though how the 2 processes are connected is unclear (van den Ameele and Brand, 2019). An additional mechanism by which NB-intrinsic genes have been demonstrated to alter the temporal gradient program is by regulating the competence of NBs to respond to extrinsic signals. The regulation of NB temporal gradient patterning by extrinsic cues is the focus of the next section.
Extrinsic regulation of Imp and Syp NB patterning
The protracted nature of the temporal patterning of postembryonic NBs allows for extrinsic signals to regulate transitions to couple neurogenesis to organismal development. The extrinsic steroid hormone ecdysone has been shown to play an important role in regulating the timing of temporal transitions in postembryonic NBs. In Type II NB lineages, ecdysone signaling is required at 60 h ALH for the transition from the early Imp/Chinmo window to the late Syp/Broad/E93 window (Syed et al., 2017; Fig. 1c). Removing ecdysone signaling by expressing a dominant-negative version of the ecdysone receptor (EcR-DN) in the NB results in the prolonged expression of Imp and Chinmo and low or no expression of the late factors Syp and E93. Whether ecdysone signaling affects each of these factors independently or acts via a single common upstream effector remains unclear. The observation that Syp is not required for Broad and E93 expression indicates that ecdysone plays a Syp independent role in activating these late genes. Ecdysone plays a general role in regulating the timing of postembryonic NB temporal transitions; in the majority of Type I NBs in the central brain, the loss of ecdysone signaling results in the failure of NBs to undergo the Imp/Chinmo to Syp/Broad/E93 transition.
What regulates the timing of the ecdysone-mediated shift from early to late gene expression? Early intrinsic Svp expression in Type II NBs primes the NB to respond to ecdysone by activating the expression of the ecdysone receptor (EcR; Syed et al., 2017; Fig. 1c). Removing svp results in the prolonged expression of the early factors Imp and Chinmo and a complete loss of the late factors Syp, Broad, and E93. Loss of svp also results in an increase in the ratio of early born to late-born neuronal fates, which is similar to the phenotype observed in a Syp loss-of-function mutant (Ren et al., 2017). The svp mutant phenotype is more severe than that observed in EcR-DN mutants (in which low levels of Syp remain), which suggests that Svp may have additional targets outside of EcR in its regulation of the early to late transition (Syed et al., 2017). In the embryo, Svp plays a similar switching role in the NBs of the ventral nerve cord where it is required for the switch from the Hunchback to Kruppel tTF windows (Kanai et al., 2005). Could Svp regulate the early to late postembryonic transition by mediating a switch between tTFs? Intriguingly, the tTF Castor is expressed in an early window in Type II NBs and its overexpression results in the delay of late gene expression, suggesting that the Castor window must be closed to allow for the early–late transition (Ren et al., 2017; Syed et al., 2017). In the future, it will be important to determine whether Svp directly activates EcR expression, or if it indirectly affects it, potentially by mediating a tTF switch.
Ecdysone signaling is also required in the mushroom body lineage where it regulates both the early and late temporal transitions (Fig. 1a). In the α’/β’ to α/β transition, ecdysone forms a negative feedback loop with Chinmo in postmitotic neurons to regulate the switch from intermediate to late fates (Wu et al., 2012). Chinmo is initially required for the expression of EcR in α’/β’ neurons (Marchetti and Tavosanis, 2017). During the prepupal stage, ecdysone signaling acts through EcR to promote the expression of the let-7 miRNA, an ecdysone inducible small regulatory RNA (Wu et al., 2012; Marchetti and Tavosanis, 2017). let-7 subsequently downregulates Chinmo via post-transcriptional control at its 3′ UTR (Wu et al., 2012). The loss of Chinmo as a result of this feedback loop allows for the transition between the α’/β’ to the pioneering α/β windows (Wu et al., 2012; Marchetti and Tavosanis, 2017). The distinct roles played by ecdysone in the temporal transitions of mushroom body neurons vs Type II NBs provides another example of how the Imp/Syp gradients and their outputs are regulated in a lineage-specific manner.
Recently, a second extrinsic signal, the Activin signaling pathway, has been shown to control temporal transitions in the mushroom body lineage (Fig. 1a). In a loss of function of the Activin receptor, Baboon, Imp levels remain high in the NB, leading to the loss of Mamo expression and the α’/β’ neuronal fate (Marchetti and Tavosanis, 2019; Rossi and Desplan, 2020). Baboon is activated by the Activin ligand Myoglianin (Myo), which is released by local glial cells in the mid-third instar larva (Awasaki et al., 2011; Upadhyay et al., 2017). The timing of Myo release thus directly regulates the timing of the γ to α’/β’ transition (Awasaki et al., 2011). Future analyses should investigate how Baboon signaling accelerates the decrease in Imp expression (Rossi and Desplan, 2020). It will also be interesting to determine whether Activin signaling plays a role in regulating temporal transitions in other postembryonic NB lineages.
Beyond diversity: Imp and Syp regulation of NB growth and termination
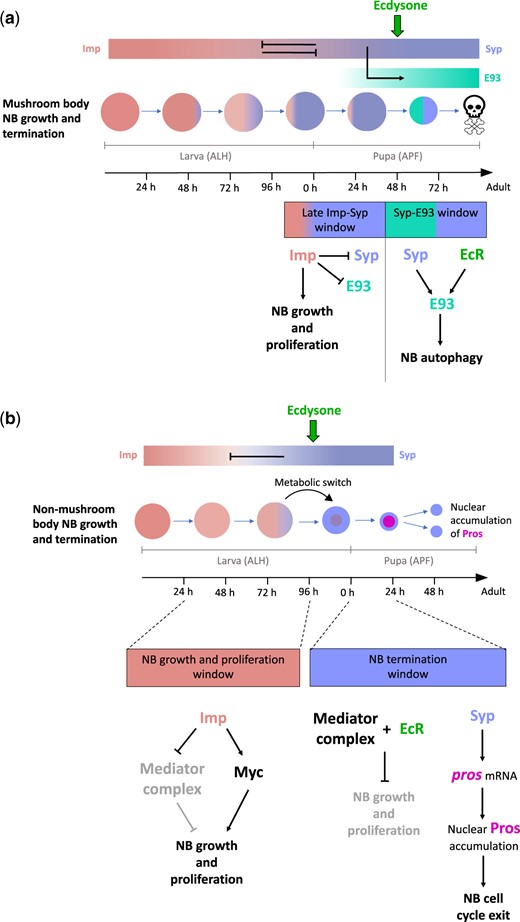
NBs undergo stage-specific termination at the end of neurogenesis via a process termed decommissioning (Yang et al., 2017). During this process, NBs slow their growth and proliferation rate before being eliminated via cell cycle exit or autophagy. Imp and Syp play critical roles in the regulation of decommissioning; loss of Syp, or overexpression of Imp, leads to a failure in NB termination and the persistence of NBs in the adult brain.
Imp prevents decommissioning by promoting NB growth and proliferation (Yang et al., 2017; Samuels, Järvelin, et al., 2020; Fig. 2). As NBs age, they undergo a switch from the use of glycolysis as an energy source to oxidative phosphorylation (Homem et al., 2014; van den Ameele and Brand, 2019). This metabolic transition, which is induced by ecdysone signaling and the mediator complex (a 30-subunit protein complex, which regulates the initiation of transcription), results in the slowing of growth and the subsequent reduction of NB size over successive divisions (Homem et al., 2014). Imp expression promotes NB growth by inhibiting members of the mediator complex (Homem et al., 2014; Yang et al., 2017). This inhibition is likely direct, as Imp binds the mRNAs of several mediator complex genes, including Med6, Med27, and Med31 (Table 1). Imp additionally promotes NB growth by directly binding to and stabilizing transcripts of the myc growth factor (Samuels, Järvelin, et al., 2020). In older NBs, Syp antagonizes Imp to promote the slowing of growth and shrinking of NBs. Syp also promotes decommissioning by stabilizing pros mRNA, leading to the accumulation of Pros in the nucleus and cell cycle exit (McDermott et al., 2014; Yang et al., 2017; Samuels, Arava, et al., 2020).
Regulation of NB growth and decommissioning. a) Imp and Syp regulate mushroom body NB growth and termination. During larval stages, Imp promotes NB growth and proliferation. In early pupal stages, Imp continues to promote NB growth and proliferation, in part by repressing Syp and E93 expression. In the mid-stage pupa, intrinsic Syp and extrinsic ecdysone signaling upregulate E93, which subsequently induces autophagy in late pupal NBs. b) Imp and Syp regulate nonmushroom body NB growth and termination. During larval stages, Imp promotes NB growth and proliferation by promoting Myc expression, and by repressing components of the mediator complex. In the prepupa, ecdysone signaling, coupled with the mediator complex, initiates a metabolic switch, which results in a decrease in NB growth. High Syp expression stabilizes pros transcripts, which results in the nuclear accumulation of Pros and cell cycle exit.
The majority of Type I and II NBs terminate neurogenesis at 24 h APF via the mechanism outlined above. The notable exception is the mushroom body NBs, which decommission much later, at 96 h APF (Yang et al., 2017; Fig. 2). How do mushroom body NBs escape decommissioning in the early pupa? The shallow complimentary Imp and Syp gradients in mushroom body NBs result in the expression of low Imp levels into pupal development (Liu et al., 2015; Yang et al., 2017). Thus, during the ecdysone and mediator complex induced shrinking of most central brain NBs at the onset of pupation, mushroom body NBs continue to regrow after each division. The eventual termination of these NBs occurs via autophagy (and not cell cycle exit) near the end of pupal development (Pahl et al., 2019). In the later stages of pupal development, Syp, together with ecdysone signaling, promotes the expression of E93. High levels of E93 activate autophagy, which results in the elimination of mushroom body NBs.
Given their role in regulating NB growth and termination during development, it is perhaps not surprising that Imp and Syp also influence the tumorigenic potential of NBs (Narbonne-Reveau et al., 2016; van den Ameele and Brand, 2019). NB-derived tumors can be generated by the manipulation of cell cycle or differentiation genes. In pros mutant NB lineages, GMCs revert to NB-like cells that can continue to proliferate (Narbonne-Reveau et al., 2016; Genovese et al., 2019). Inducing pros mutations in young NBs, when Imp levels are high, results in the formation of large, metastatic tumors that persist into the adult. In contrast, generating pros mutations in older NBs, when Imp is absent, results in only a few extra NB divisions (Narbonne-Reveau et al., 2016; Genovese et al., 2019). Future studies should determine if the regulatory relationships between early and late temporal factors that have been elucidated during normal postembryonic NB development are conserved in tumorigenic NBs.
Concluding remarks
During the postembryonic development of the fly CNS, opposing temporal gradients of the RNA-binding proteins Imp and Syp regulate neuronal fate specification through a hierarchical gene regulatory network (Ren et al., 2017; Syed et al., 2017; Liu et al., 2015, 2019). Intrinsic and extrinsic regulators act on this network to both increase neuronal diversity and regulate the timing of temporal transitions (Marchetti and Tavosanis, 2017, 2019; Ren et al., 2017; Syed et al., 2017; Rossi and Desplan, 2020). By controlling the length of temporal windows and the timing of lineage termination, Imp and Syp also regulate the relative number of each neuronal cell type that is generated in a lineage.
Imp and Syp temporal gradients may also control circuit assembly by coupling birth order to neuronal morphology. In central brain Type I lineages, NBs generate neurons with decreasing morphological complexity over time (Lee et al., 2020). Early neurons, with extensive and elaborate projections, are likely born in a high Imp temporal window and may serve a pioneering function required for the initial establishment of connectivity in the circuit. It will be interesting to determine whether these neurons are lost in an Imp mutant background, and if the absence of their pioneering arborizations results in defects in circuit architecture. Imp and Syp temporal gradients may also couple birth order with circuit assembly in the postembryonic lineages of the ventral nerve cord. The birth order of motor neurons in the NB 2-3 lineage is correlated with their muscle innervation pattern along the proximal-distal axis of the femur (Brierley et al., 2012; Baek et al., 2013). The region-specific innervation pattern of these neurons is regulated by temporal gradients of the transcription factor Antennapedia (Antp); higher Antp levels in early-born neurons promote more proximal targeting, whereas lower levels promote more distal targeting (Baek et al., 2013). An intriguing possibility is that Imp and Syp post-transcriptionally regulate Antp transcripts to control the birth-order-dependent proximal-distal innervation patterns of these neurons.
Future analyses should also investigate the degree by which the temporal gradient patterning mechanisms uncovered in the fly are conserved in vertebrate neurogenesis. Indeed, the homologs of Imp, let-7, and Svp appear to play conserved roles in the temporal patterning of vertebrate NSCs. In mice, IMP1, the vertebrate ortholog of Imp, is highly expressed in neural progenitors during early embryonic stages and rapidly declines in expression by late fetal development (Nishino et al., 2013). IMP1 is required for NSC maintenance and its loss results in a reduction in brain size. Let-7 functions as a developmental switch that regulates age-related changes within several organisms (Pasquinelli et al., 2000; Reinhart et al., 2000; Toledano et al., 2012). In the mouse, increased Let-7 expression at the onset of postnatal development represses IMP1 in neural progenitors and mediates progenitor cell cycle exit (Nishino et al., 2013). Finally, the vertebrate homologs of Svp, Chick Ovalbumin Upstream Promoter-Transcription Factors (COUP-TFI and II), are expressed early in neural progenitors in the developing mouse brain, where they are required for the switch between neuronal and glial production (Naka et al., 2008). The role of the COUP genes is thus similar to that of Svp, which regulates the switch from early born to late-born neurons in Type I and II NB lineages (Ren et al., 2017). As additional temporal gradient patterning genes are identified in the fly, it will be interesting to determine if they play conserved roles in the developing vertebrate CNS.
Funding
This work was supported by an NSERC Discovery Grant (RGPIN-2015-06457).
Conflicts of interest
None declared.