-
PDF
- Split View
-
Views
-
Cite
Cite
Markus Tögel, Günther Pass, Achim Paululat, Wing hearts in four-winged Ultrabithorax-mutant flies—the role of Hox genes in wing heart specification, Genetics, Volume 220, Issue 1, January 2022, iyab191, https://doi.org/10.1093/genetics/iyab191
Close - Share Icon Share
Abstract
Wings are probably the most advanced evolutionary novelty in insects. In the fruit fly Drosophila melanogaster, proper development of wings requires the activity of so-called wing hearts located in the scutellum of the thorax. Immediately after the imaginal ecdysis, these accessory circulatory organs remove hemolymph and apoptotic epidermal cells from the premature wings through their pumping action. This clearing process is essential for the formation of functional wing blades. Mutant flies that lack intact wing hearts are flightless and display malformed wings. The embryonic wing heart progenitors originate from two adjacent parasegments corresponding to the later second and third thoracic segments. However, adult dipterian flies harbor only one pair of wings and only one pair of associated wing hearts in the second thoracic segment. Here we show that the specification of WHPs depends on the regulatory activity of the Hox gene Ultrabithorax. Furthermore, we analyzed the development of wing hearts in the famous four-winged Ultrabithorax (Ubx) mutant, which was first discovered by Ed Lewis in the 1970s. In these flies, the third thoracic segment is homeotically transformed into a second thoracic segment resulting in a second pair of wings instead of the club-shaped halteres. We show that a second pair of functional wing hearts is formed in the transformed third thoracic segment and that all wing hearts originate from the wild-type population of wing heart progenitor cells.
Introduction
The enormous success of insects is undoubtedly rooted in the most astonishing evolutionary novelty: the origin of powered flight. How exactly wings developed in the course of evolution is still subject of an old and fierce controversy mostly concerned with the evolution of the cuticular wing blade (Niwa et al. 2010; Clark-Hachtel and Tomoyasu 2016; Alexander 2018). However, active flight requires much more than just external wings. It needs a powerful musculature, innervation, tracheal supply, and also circulation. The latter is facilitated by accessory pulsatile organs called wing circulatory organs or wing hearts, which are located in the scutellum, a small dorsal elevation on each wing bearing segment (Pass et al. 2015; Figure 1, A–D). In Drosophila, and probably also in all other insects, wing hearts play an essential role in a process called wing maturation (Tögel et al. 2008). Insects unfold their wings by pumping hemolymph into the space between the dorsal and ventral wing surface (Robertson 1936; Johnson and Milner 1987). The epidermal cells that until then connected the dorsal and ventral wing surfaces undergo apoptosis and delaminate from the cuticle (Kimura et al. 2004; Kiger et al. 2007; Thuma et al. 2018). In Drosophila, the generated cell debris and excess hemolymph are removed by the sucking action of the wing hearts. In addition, the dorsal and ventral wing surfaces are brought closely together so that integrin-mediated bonding can occur (Brower and Jaffe 1989). This process takes about 2–3 h and is called wing clearance because the wings gradually change their appearance from opaque (freshly unfolded) to transparent (mature). Flies with nonfunctional or lacking wing hearts fail to undergo wing clearance and are unable to fly (Tögel et al. 2008, 2013a). After wing maturation, wing hearts are crucial for maintaining the biomechanical properties of the wing blade cuticle through continuous hydration and the metabolic supply of the numerous sensory organs located on these body appendages (Pass 2018).
Morphology and development of wing hearts. (A) Dorsolateral view of an adult thorax showing the location of the scutellum in T2. The scutellar arm connects the scutellum to the posterior wing vein. (B) Epifluorescence image of a pharate adult with bilaterally located wing hearts in the scutellum (dorsal view). (C) Lateral view of a wing heart located in the lateral corner of the scutellum. (D) cLSM image of a wing heart expressing handC-GFP (lateral view). (E) Schematic representation of stages 13 and 17 embryos illustrating the distribution of EPCs along the anterior–posterior axis. The prospective WHPs are depicted in green, all other EPCs in gray. WHPs lose Tinman expression and relocate in front of the heart. The EPCs anterior to the WHPs remain close to the heart and form the OHS together with the EPCs in A1 (PS6). (F) Schematic of postembryonic wing heart development. WHPs proliferate and undergo passive relocation as well as active migration before their final differentiation into mature wing hearts. Numbers indicate cells originating from the four pairs of WHPs (A1, first abdominal segment; EPC, Even-skipped positive pericardial cell; OHS, outflow hanging structure; sc, scutellum; T2, second thoracic segment; T3, third thoracic segment; wh, wing heart; cartoons of developmental stages adapted from Hartenstein 1993).
The single pair of wing hearts in the adult Drosophila mesothoracic segment (T2; Figure 1, B–D) originates from eight embryonic wing heart progenitors (WHPs) located in two pairs on either side of the embryo in parasegments 4 and 5 (PS4 and PS5; Figure 1E). After their specification in the embryo, WHPs remain dormant until the transition from the second to the third larval instar upon which they initiate proliferation. Although originating from two different PS, all WHPs merge during the prepupal stage into one single large cell cluster. This is achieved through passive relocation and occurs as part of the overall remodeling of the prepupa due to the eversion of the imaginal discs. During the subsequent pupal stages, the cluster splits and the cells actively migrate to the lateral corners of the forming scutellum, where they differentiate into the muscular wing hearts (Tögel et al. 2008, 2013b; Lehmacher et al. 2009; Figure 1F).
WHPs are a subpopulation of the well-characterized Even-skipped positive pericardial cells (EPCs; Carmena et al. 2002; Park et al. 1998; Su et al. 1999; Han et al. 2002; Han and Bodmer 2003), which arise in pairs in each PS from PS2 to PS12 and as a single EPC in PS14 on either side of the embryo (Figures 1E and 7). All EPCs are interconnected by cell protrusions that extend between neighboring EPCs on the same side of the embryo and after dorsal closure also between contralateral EPCs (Tögel et al. 2008; Zmojdzian et al. 2018). At least in WHPs, these connections persist in post-embryonic stages and may be important for their passive relocation during the prepupal stage.
Initially, EPCs express Even-skipped (Eve) protein, Tinman (Tin) protein, and Hand (visualized by the handC-GFP reporter). Later in embryonic development, they also express and secrete Pericardin, a collagen IV-like structural protein that is incorporated into the cardiac matrix by the Lonely heart adapter protein (Drechsler et al. 2013; Rotstein et al. 2018; Wilmes et al. 2018), which makes EPCs important contributors to the structural integrity of the larval cardiac tube.
WHPs are not the only EPCs that differentiate into functional post-embryonic anatomical structures. It has been shown recently that the EPCs in PS3, PS4, and PS6 contribute to the anterior suspension of the cardiac tube by forming the so-called outflow hanging structure (OHS; Figure 1E), while the posterior EPCs along the heart proper form an actin-rich network around the heart reminiscent of an epicardium (Zmojdzian et al. 2018; Figure 1E).
So far, the only known difference between WHPs and the remaining EPCs is the downregulation of Tinman expression in WHPs from stage 13/14 onward and the continued activation of the handC-GFP reporter in post-embryonic stages (Tögel et al. 2008; Pass et al. 2015). This is supported by the findings that both the ectopic expression of tinman or the loss of Hand in WHPs inhibits wing heart differentiation (Tögel et al. 2008, 2013a, 2013b).
Ever since our initial study on the developmental origin of wing hearts, we were intrigued by the fact that WHPs arise in both thoracic segments T2 (PS4) and T3 (PS5), even though the dipteran lineage has lost functional wings on T3. Could this represent the ancestral state dating back to a time when dipterans still had four wings like most other insects? And how is this regulated? The fact that EPCs are distributed along the anterior–posterior axis and WHPs are specified within parasegmental boundaries, suggested an involvement of homeotic selector genes (Hox) in this process. Indeed, Hox genes have been found to control the diversification of EPCs along the anterior–posterior axis (Zmojdzian et al. 2018). This prompted us to focus on two main questions: (1) How are the anterior and posterior boundaries specified that delimit the loss of Tinman within the row of EPCs and thus control the number of WHPs? (2) What happens if both T2 and T3 bear wings (four-winged fly) like in an ancestral condition? Would this lead to functional wing hearts in both thoracic segments and how would their development differ from that of a “modern” fly? To address these questions, we studied Hox gene expression in WHPs and their specification in several Hox null mutant backgrounds. In addition, we generated four-winged flies to investigate wing heart development in animals carrying wings on both the meso-and metathoracic segment. Our data revealed that Ubx controls the posterior boundary of the WHP field. Furthermore, we could show that the eight WHPs in PS4 and PS5 are sufficient to build two pairs of functional wing hearts in four-winged flies. This indicates that the specification of WHPs in two adjacent PS most likely reflects the ancestral evolutionary state of the dipteran lineage with the differentiation of forewings and hindwings in T2 and T3.
Materials and methods
Fly stocks
The triple mutant y, w; abx, bx3, pbx/TM6B, Tb, which causes the homeotic transformation into four-winged flies, was a gift from Ernesto Sanchez-Herrero. The history of its discovery has been described by Lewis (2007). All handC-GFP reporter lines have been described previously (Sellin et al. 2006). The handCΔ395-633-GFP line harbors a 239 bp deletion in the handC enhancer fragment and drives expression exclusively in embryonic tinman positive cardioblasts and all EPCs (unpublished data). The following lines were obtained from the Bloomington Drosophila Stock Center (BDSC) at Indiana University: BL2185 Scr2, first described as EdK6 (Lewis et al. 1980a); BL3020 Antp25, first described as EfW10 (Lewis et al. 1980b); BL3474 Ubx9.22 (Kerridge and Morata 1982). The TM2, Ubx130 balancer chromosome was introduced by Lewis (1952) and maintained in our laboratory as TM2, Ubx130/TM6B, Tb1. The Ubx130 allele is caused by a breakpoint within the Ubx locus that has not been molecularly characterized so far. Ubx130 heterozygous animals show an enlarged distal segment of the haltere, while homozygous Ubx130 animals are lethal (Lewis 1952). The third chromosomal balancer TM3, Dfd-lacZ, Sb1 (TDLZ, Sb), used to distinguish between hetero- and homozygous mutant embryos, was a gift from Alan Michelson.
Immunohistochemistry
Embryos were fixed as described previously (Sellin et al. 2009) and stored in 100% ethanol. Prior to the staining, embryos were washed 3–4 times in PBT (phosphate buffered saline supplemented with 0.1% Tween) for about 5–10 min each. Primary antibodies were applied overnight at 4°C. Embryos were then washed 5 times in PBT for 5 min each and subsequently blocked for 30 min in Rotiblock (Roth, Germany) diluted 1:10. Secondary antibodies were applied for 2–3 h at room temperature or overnight at 4°C. After three washes in PBT for about 5–10 min each, embryos were mounted in Fluoromount G (Southern Biotech). All washes, blocking, and incubation with secondary antibodies were performed with gentle agitation. Primary and secondary antibodies as well as the blocking solution were diluted in PBS.
The following primary antibodies were used in this study: mouse anti-Antp 1:3 (8C11 or 4C3, supernatant, DSHB), mouse anti-Even-skipped 1:2 (2B8, supernatant, DSHB), rabbit anti-GFP 1:500 - 1:1000 (ab6556, Abcam), mouse anti-GFP 1:1000 (A11120, Invitrogen), mouse anti-Scr 1:3 (6H4.1, supernatant, DSHB), rabbit anti-Tin 1:500 (Bodmer 1993), mouse anti-Ubx/Abd-A 1:3 (FP6.87, supernatant, DSHB), mouse anti-β-Galactosidase 1:2000 (Z3781 or Z3783, Promega) and rabbit anti-β-Galactosidase 1:4000 (Cappel number 55976, MP Biomedicals). The Ubx antibody FP6.87 recognizes all isoforms of the Ultrabithorax and Abdominal-A proteins (Kelsh et al. 1994), a feature, which was important to us to detect all Ubx protein being present in WHPs or the adjacent epidermis. Secondary antibodies used in this study were anti-rabbit or anti-mouse conjugated Cy2 1:100 (Dianova) and anti-rabbit or anti-mouse conjugated Cy3 1:200 (Dianova). The monoclonal antibodies 8C11, 4C3, 6H4.1, FP6.87, and 2B8 developed by Brower, D. (University of Arizona), White, R. (University of Cambridge), and Zinn, K. (California Institute of Technology) were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242.
Microscopy and image processing
Larvae, prepupae, and pupae were prepared for imaging as described previously (Tögel et al. 2008, 2013b; Tögel 2012). For the removal of the puparium, pupae were placed ventral side down onto a double-sided Scotch tape affixed to a microscope slide. Then the operculum was carefully removed with fine forceps and a longitudinal incision was made on one side of the puparium starting at the posterior edge of the operculum all the way to the posterior end of the puparium. This was achieved by repeatedly inserting one tip of a forceps between pupa and puparium and then pulling up to break open the puparium. Finally, each side of the puparium was bent open and adhered to the Scotch tape to free the pupa. Epifluorescence images were acquired on a Leica MZ16FA stereomicroscope. Confocal microscopy was performed on a Zeiss Pascal 5 confocal laser scanning microscope equipped with a Plan-Neofluar 25x/0.8 objective. All confocal images were pre-processed with the Zeiss software and represent maximum intensity projections of image stacks. Images were edited with Affinity Photo and assembled into figures with Affinity Designer (Serife).
Results
Expression of Hox genes in WHPs
WHPs are a subpopulation of EPCs and as such are considered to belong to the cardiac mesoderm. Both the Hox gene expression in the cardiac tube as well as in EPCs has been studied with antibody stainings in late stage embryos (roughly stage 16). The expression in the cardiac tube can be summarized as follows: No Hox gene expression in the anterior-most part. Antp protein expression is weak in T3, strong in A1, and weak in A2. Ubx protein is weak in A2, strong in A3 and A4, and weak in A5 (barely detectable in A6–A8). Abd-A protein defines the heart proper and thus is weak in A5, strong in A6 and A7, and weak in A8. Abd-B protein expression is weak in A7 and strong in the last four cardioblasts constituting the end of the heart in A8 (Lo et al. 2002; Lovato et al. 2002; Ponzielli et al. 2002; Perrin et al. 2004). The Hox gene expression in EPCs generally resembles the pattern in the cardiac tube: The EPCs in the region of the anterior aorta (originating from PS2 and PS3) do not express any Hox genes. Antp protein expression is found in the WHPs (originating from PS4 and PS5), in the EPCs in A1 (PS6) and at a lower level in the EPCs within A2 to A4 (PS7 to PS9). Ubx protein is expressed in the EPCs in A3 and A4 (PS8 and PS9) while Abd-A protein is detected in EPCs next to the heart proper in A5 to A7 (PS10 to PS12). Abd-B protein is found in the two EPCs close to the posterior end of the heart in A8 (PS14; Zmojdzian et al. 2018).
However, WHPs arise much earlier in development and Hox genes can not only act cell autonomously, but also through signaling from neighboring tissues or germ layers (Miller et al. 2001). We therefore investigated Hox gene expression in the dorsal mesoderm at stage 13/14 right when the EPCs in PS4 and PS5 lose Tinman protein expression and become WHPs (Figure 2). For comparison, we also investigated the expression at stage 16/17 after WHP relocation in front of the heart (Figure 3). Based on their published expression patterns, only Scr, Antp and Ubx have the potential to be critical in WHP specification: Scr protein is expressed in the epidermis of the labial segment (PS2) and T1 (PS3) as well as in the mesoderm of PS3 in somatic muscles and around the anterior midgut from stage 11 onward (Mahaffey and Kaufman 1987; Martinez-Arias et al. 1987; Riley et al. 1987). Antp protein expression is found in the epidermis in the three thoracic segments T1 (PS3) to T3 (PS5) at stage 13 (Carroll et al. 1986; Martinez-Arias 1986; Wirz et al. 1986) and is detected in the mesoderm of T3 (PS5) in myoblasts and in a region of the visceral mesoderm (Martinez-Arias 1986). Ubx protein is expressed in the epidermis from PS6 to PS12 at stage 13 and in a similar pattern in the somatic mesoderm although slightly shifted posteriorly by one PS. In the visceral mesoderm, Ubx protein expression is found around the developing midgut (Akam and Martinez-Arias 1985; see also Figure 7).
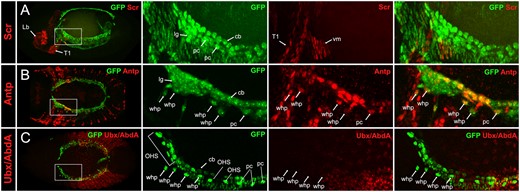
Expression of Hox genes in embryonic WHPs, stage 13/14. cLSM images of stage 13/14 handC-GFP or handCΔ395–633-GFP embryos stained for GFP and for anti-Scr (A), anti-Antp (B), or anti-Ubx/Abd-A (C). Left panel shows a dorsal overview of the stained embryo. Panels to the right show an enlargement of the region of WHP specification (position is indicated by dashed lines in the overview). WHPs express only Antp at this stage. cb, cardioblast; Lb, labial segment; lg, lymph gland; OHS, EPCs forming the outflow hanging structure, pc, pericardial cell; T1, first thoracic segment; vm, visceral mesoderm; whp, wing heart progenitor. handC-GFP and handCΔ395–633-GFP reporter lines are explained in the Materials and Methods section.
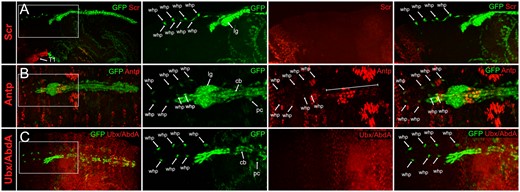
Expression of Hox genes in embryonic WHPs, stage 16/17. cLSM images of stage 16/17 handC-GFP or handCΔ395–633-GFP (Sellin et al. 2006) embryos stained for GFP and anti-Scr (A, lateral view), anti-Antp (B, dorsal view), or anti-Ubx/Abd-A (C, dorsal view). Left panel shows an overview of the stained embryo. Panels to the right show an enlargement of the WHPs located in front of the heart (position is indicated by dashed lines in the overview). WHPs (marked by arrows) only express Antp at this stage. The bracket indicates Antp expression in the heart. lg, lymph gland; cb, cardioblast; pc, pericardial cell; whp, wing heart progenitor. handC-GFP and handCΔ395–633-GFP reporter lines are explained in the materials and methods section.
To visualize the cardiac mesoderm (heart, pericardial cells, and lymph glands) in the expression analyses, we used our handC-GFP reporter lines (Sellin et al. 2006). We found that Scr protein is not expressed in any cells of the cardiac mesoderm including the WHPs, neither at stage 13/14 nor at stage 16/17 (Figures 2A and 3A). These findings confirm the previously reported lack of expression in the developing heart (Lo et al. 2002; Perrin et al. 2004).
In contrast, Antp protein is expressed in consecutive cardioblasts and in several pericardial cells at stage 13/14. Based on their position, the Antp positive pericardial cells next to the lymph glands are the relocating WHPs (Figure 2B). At stage 16/17, Antp was still detected in the WHPs, now located in front of the heart, suggesting that the expression pattern established at stage 13/14 is maintained until the end of embryogenesis (Figure 3B). This is also the case with the cardiac expression pattern. At stage 16/17, it consists of three to four weakly stained pairs of consecutive cardioblasts in T3, immediately followed by four strongly stained pairs of cardioblasts in segment A1, and another set of four weakly stained pairs in segment A2 (Lo et al. 2002). A very similar pattern was already observed at stage 13/14 (Figures 2B and 3B). Other pericardial cells in the adjacent posterior segments were also Antp positive but could not be identified as EPCs due to broad expression of the handC-GFP reporter line.
For the Ubx staining an antibody was used that recognizes all isoforms of both the Ultrabithorax and Abdominal-A proteins (Kelsh et al. 1994). This resulted in a very strong staining in the posterior epidermis which made the expression in the underlying heart difficult to see. Nevertheless, a faint expression was detectable in some cardioblasts in A4, A5, and A6 in accordance with the previously published expression studies (Lo et al. 2002). The WHPs, however, reside in the segments anterior to the region of strong epidermal expression and could easily be analyzed. To be able to investigate also the expression in the EPCs adjacent to the WHPs, we used the handCΔ395-633-GFP reporter line which is only active in tinman positive cardioblasts and EPCs. At both stages (13/14 and 16/17) no expression could be detected in WHPs or the EPCs in A1 and A2 (Figures 2C and 3C). This is consistent with the published expression pattern of stage 16 EPCs in these segments (Zmojdzian et al. 2018). Again, the expression pattern established at stage 13/14 was maintained until the end of embryogenesis.
Ubx defines the posterior border of WHP specification
To investigate the requirements for any of the three Hox genes Scr, Antp, and Ubx during WHP specification, we utilized well-established null mutant alleles: Scr2 (Lewis et al. 1980a) harbors a point mutation that results in a premature stop codon. The truncated protein lacks several conserved Scr protein domains including the homeodomain (Sivanantharajah and Percival-Smith 2009). Antp25 (Lewis et al. 1980b) has not been molecularly characterized but is considered a null allele based on genetic evidence (Wakimoto and Kaufman 1981). Ubx9.22 (Kerridge and Morata 1982) carries a deletion spanning the splice acceptor site of the 3’ exon and 48 codons of the homeodomain. The resulting protein is most likely splice defective and out of frame downstream of the truncated homeodomain (Weinzierl et al. 1987). All three alleles are recessive lethal with animals dying at the end of embryogenesis or shortly thereafter (Wakimoto and Kaufman 1981; Kerridge and Morata 1982; Sivanantharajah and Percival-Smith 2009).
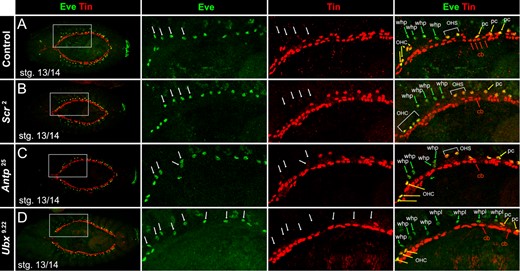
The loss of Tinman protein expression in the EPCs of PS4 and PS5 at stage 13/14 is a hallmark of WHP specification (Figure 4A). It is accompanied by the relocation of the WHPs in front of the heart, while all other EPCs (especially the ones in PS2 and PS3) remain close to the heart (Tögel et al. 2008; Zmojdzian et al. 2018). We therefore focused our analysis on this developmental stage and investigated Tinman protein expression in EPCs in various mutant backgrounds. Like in the control, the EPCs originating in PS2 and PS3 remain close to the heart and the WHPs in PS4 and PS5 start their relocation in front of the heart in all three mutants. In addition, the anterior border of WHP specification was not affected in any of the mutants. Neither loss of Scr nor Antp gene function led to an expansion of Tinman protein downregulation in anterior direction. In both the Scr2 and Antp25 mutants, the wild-typic number of eight WHPs was observed (Figure 4, B and C). However, in Ubx9.22 mutants, the posterior border was shifted by two PS toward the abdomen. This increased the total number of EPCs with downregulated Tinman protein expression to 16. Based on our studies in the wild type, this would suggest that additional WHPs in the non-wing-bearing first two abdominal segments have been specified (Figure 4D). However, Ubx9.22 mutants do not survive past embryonic stages which make it impossible to determine the actual fate of these cells. We will therefore refer to these additional Tinman protein negative EPCs as WHP-like cells.
WHP specification in Hox mutant background, stage 13/14. cLSM images of control and mutant embryos stained for anti-Eve and anti-Tin. Left panel shows a dorsal overview of the stained embryo. Panels to the right show an enlargement of the region of WHP specification (position is indicated by dashed lines in the overview). (A) In the control (Scr2/TDLZ, Sb) as well as in the Scr2 (B) and the Antp25 (C) null mutant, the EPCs in PS4 and PS5 downregulate Tinman protein and become the four WHPs (arrows) on either side of the embryo. (D) In Ubx9.22 null mutants, eight EPCs downregulate Tinman protein on either side of the embryo. These consist of the four WHPs in PS4 and PS5 and four WHP-like cells in PS6 and PS7. cb, cardioblast; OHS, EPCs forming the outflow hanging structure; pc, pericardial cell; whp, wing heart progenitor; whpl, wing heart progenitor-like.
These findings show that Antp, although expressed in WHPs, is not required for downregulation of Tinman protein during WHP specification. On the other hand, Ubx, which is not expressed in WHPs and the EPCs in A1 and A2, is necessary to restrict WHP specification to the thorax by controlling Tinman protein expression in the posterior EPCs, likely through instructive signals from the overlying epidermis.
Generating four-winged flies
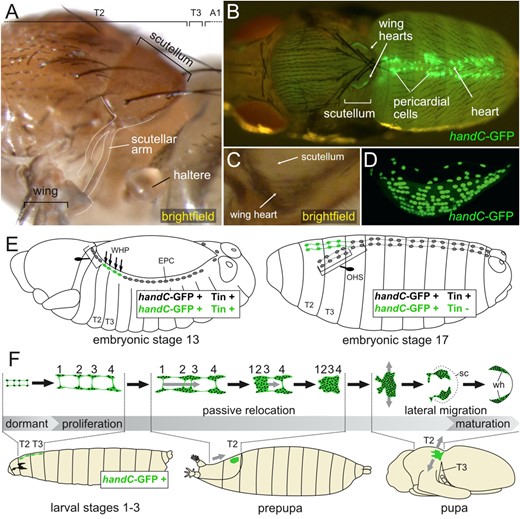
To address the question of what happens during wing heart development when both T2 and T3 bear wings as well as to determine the fate of the WHP-like cells which are specified in A1 and A2 of Ubx null mutants, we generated four-winged flies. In the original work by Lewis, a complete transformation of T3 to T2 is seen in the homozygous triple mutant anterobithorax (abx), bithorax (bx3), postbithorax (pbx; Lewis 1978). The same observation was made by Dutta and colleagues (Dutta et al. 2010). Unfortunately, homozygous triple mutants of our Drosophila stock with the genotype y, w; abx, bx3, pbx/TM6B, Tb did not survive until adult stages for unknown reasons. Instead, we were able to obtain four-winged flies in heterozygous animals carrying the triple mutant chromosome anterobithorax (abx), bithorax (bx3), postbithorax (pbx) over the balancer TM2, Ubx130. This balancer chromosome harbors chromosomal rearrangements with breakpoints in the 89D and 89E region, causing a mutation within the Ubx locus (Lewis 1952; FlyBase: Drysdale et al. 2005; Figure 5, A and B). The precise nature of this mutation is still unknown, since the breakpoints have not been molecularly characterized. All animals with the genotype abx, bx3, pbx/TM2, Ubx130 display a transformation of T3 to T2, with wings instead of halteres and a notum with a scutellum. However, the left and right halves of the notum often do not meet at the midline. This results in a homeotically transformed metathorax (HT2) that may exhibit a dorsal cleft (Figure 5C). Such a split thorax occurred in our study in about 55% of all HT2. Other frequently found phenotypes were buckled bristles on the scutellum of T2 and malformed legs on HT2. In accordance with earlier observations by Lewis, flies with four wings have difficulties eclosing and often need manual assistance. Most of the times, they fail to wriggle themselves through the opening of the operculum and die within the puparium. Others are unable to free their legs and remain attached to the puparium. Nevertheless, by removing the puparium manually with forceps, it is possible to rescue four-winged flies. Such flies often have difficulties walking properly due to the malformed legs on HT2 and frequently end up lying on their backs. Since flies actively search for a safe suitable perch before they unfold their wings (Peabody et al. 2009), which is hindered in four-winged flies, perfectly unfolded wings are rarely observed. Moreover, the development of the indirect flight musculature is impaired in four-winged flies (Fernandes et al. 1994), resulting in inability to move the wings in a controlled manner.
Cytological location of Ubx and its homeotic mutations. (A) Ubx maps to the cytological band 89D. The homeotic mutations abx, bx3, and pbx are located in the third intron and upstream of the Ubx gene and affect cis regulatory elements [based on FlyBase (Drysdale et al. 2005) and adapted from Maeda and Karch (2006)]. (B) Translocations and inversions induced by irradiation led to a chromosomal breakpoint between band 89D and 89E within the balancer chromosome TM2, Ubx (FlyBase: Drysdale et al. 2005). (C) Flies carrying the triple mutation over the TM2, Ubx balancer display a homeotic transformation of T3 to T2 indicated by the presence of additional wings and a second scutellum. The transformed metathorax (HT2) shows in about 50% of specimens a dorsal cleft named split thorax. The formation of split thoraces is temperature independent. (D) Control and four-winged fly carrying a handC-GFP reporter. In the control (left), the mesothorax is the only large wing bearing segment. At 50 h APF, the wing hearts are visible as bilateral organs in T2 of the pupa. Adult four-winged flies (right) display two large wing bearing segments in the thorax due to the homeotic transformation of T3 to T2 (HT2). The wings are not as perfectly clear as in the control but still show signs of wing maturation. In 50 h APF pupae of completely transformed four-winged flies, two pairs of wing hearts are visible in T2 and HT2. d AH, days after hatching; h APF, hours after puparium formation; T2, second thoracic segment; T3, third thoracic segment; wh, wing heart.
Four-winged flies have two pairs of functional wing hearts
The famous picture of one of Lewis’ flies, which appeared on the cover of Science in 1983 (Lewis 1983; Lipshitz 2004), showed an animal with a nearly perfectly developed second pair of wings on a duplicated second thoracic segment. Since proper wing clearance requires functional wing hearts (Tögel et al. 2008), this image of a four-winged fly already suggested that HT2 contains a functional second pair of wing hearts. To properly investigate the presence of wing hearts in HT2 and their function, we used handC-GFP as live cell marker and analyzed late pupal stages of four-winged flies after removal of the puparium. Indeed, we found additional wing hearts in HT2 in all animals with a complete transformation (Figure 5C). To assess their functionality, we monitored these extra wing hearts over a period of time. We observed beating of all four wing hearts in four-winged flies, indicating that they are functional. Wing hearts consist of 6–8 syncytial muscles cells (Lehmacher et al. 2009), which are responsible for the contractions. Our observation of beating wing hearts in HT2 indicates that myogenesis and motoneuron innervation (Tögel et al. 2013b) also occurs in the extra wing hearts. In addition to wing heart beating, wing clearance can be used as indicator of wing heart functionality. As mentioned above, many of the four-winged flies have severe difficulties eclosing or do not unfold their wings properly. All animals that unfolded their wings were analyzed for correct wing maturation. As nonfunctional or missing wing hearts result in opaque wing blades with large blisters, the appearance of largely transparent clear wing blades indicate the presence of functional wing hearts (Tögel et al. 2008). We indeed found four-winged flies with four clear and blister-free wings, confirming that wing hearts can reach full functionality in four-winged individuals (Figure 5D).
Origin of extra wing hearts in four-winged flies
WHPs are specified in the meso- and metathoracic segments of the Drosophila embryo. However, Drosophila, like all dipteran insects, possesses only one pair of wings on the mesothorax. In the wild type, this situation is resolved by uniting all WHPs from both thoracic segments in the mesothorax during the prepupal stage. During the subsequent pupal stage, the single WHP cluster splits and the cells migrate toward the lateral corners of the scutellum to form a pair of wing hearts in T2 (Tögel et al. 2008, 2013b; see also Figure 1). However, in homozygous Ubx9.22 null mutants 16 EPCs lose Tinman expression resulting in the specification of 8 WHPs (in T2 and T3) and 8 WHP-like cells (in A1 and A2; Figure 4). Based on this observation, we considered two possible explanations for the development of an extra pair of wing hearts in four-winged flies: (1) The development follows the wild-type pattern and the extra wing hearts originate from the abdominal WHP-like cells. Similar to the recruitment of WHPs from T3 to T2, HT2 could recruit WHP-like cells from the abdomen. (2) The development deviates from the wild-type pattern and the WHPs remain in their respective segments to form wing hearts locally. In this scenario, the wing hearts in T2 would originate from WHPs in T2 and the wing hearts in HT2 from WHPs in HT2. The fate of the WHP-like cells would be unknown.
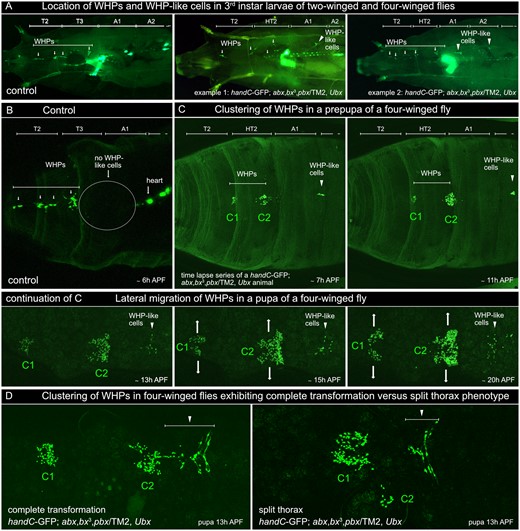
To test these hypotheses, we first analyzed, whether additional abdominal WHP-like cells are also present in 3rd instar wandering larvae and pupae of four-winged flies. Indeed, like in the Ubx null mutants, we found additional handC-GFP expressing cells in A1 and in A2 (Figure 6). These extra abdominal WHP-like cells were associated with the interconnection of the dorsal tracheal branches like all other WHPs (Tögel et al. 2013b). To study the mode of wing heart formation in more detail, we used our established live cell imaging technique to follow the WHPs from prepupal to adult stages (Figure 6, B and C). We found that in Ubx mutants with a homeotic T3 to HT2 transformation, the WHPs did not relocate into one large cluster but remained in their respective segments forming two separate smaller clusters instead. The additional abdominal WHP-like cells also remained in their abdominal segments (Figure 6C). The subsequent development followed the wild-type pattern. Each cluster split and the cells migrated into the lateral corners of the forming scutella. However, in flies were the homeotic HT2 transformation was accompanied by a split thorax phenotype, WHPs often relocated nearly completely into one single larger cell cluster, resembling the wild-type situation. This resulted in many cases in the formation of only a single wing heart on one side in HT2 (Figure 6D) or, very rarely, in a complete lack of wing hearts in HT2. These findings demonstrate that proper wing heart formation requires an intact notum and scutellum.
Wing heart development in four-winged flies. (A) Dorsal view of 3rd instar larvae of wild-type (control) and four-winged flies carrying a handC-GFP reporter. In the control, WHPs (associated with dorsal tracheal branches) are only present in T2 and T3 (arrows). Larvae of four-winged flies show additional WHP-like cells (also associated with dorsal tracheal branches) in A1 and in A2 (arrowheads). (B) cLSM image of a control prepupa (dorsal view of the region of the operculum) illustrating the lack of WHP-like cells in A1 and A2. (C) cLSM images taken from a time lapse series showing wing heart development in a four-winged fly with complete HT2 transformation. During prepupal stages, WHPs relocate into two separate clusters (C1 and C2). The WHP-like cells remain in A1 and do not merge with these clusters. During pupal stages, the WHPs in C1 and C2 start to migrate laterally (arrows) and will differentiate into mature wing hearts. The WHP-like cells still remain in A1 and do not contribute to the formation of the wing hearts. (D) Comparison of WHP cluster formation in a four-winged fly with complete transformation (left) and a four-winged fly exhibiting a split thorax phenotype (right). In the latter case, WHPs are almost completely relocated into a single cluster and only a few WHPs are found on one side in HT2. A1, first abdominal segment; A2, second abdominal segment; h APF, hours after puparium formation; HT2, homeotically transformed third thoracic segment; T2, second thoracic segment; T3, third thoracicsegment; WHP, wing heart progenitor.
Discussion
Boundaries of WHP specification
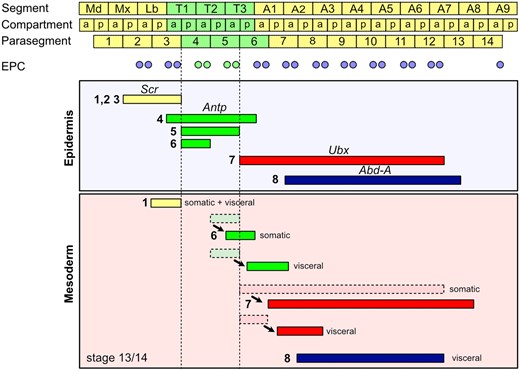
Wing hearts are genuine adult structures that develop from embryonic WHPs. The progenitors are specified as subpopulation within the so-called EPCs; Figure 1). EPCs themselves arise in pairs in each parasegment (PS) from PS2 to 12 and as a single cell in PS14 (Figure 7). Thus, EPCs are distributed in rows along the anterior–posterior axis on either side of the embryo. Initially, they express Even-skipped protein, Hand, and Tinman protein. However, the EPCs in PS4 and 5 downregulate Tinman protein expression at stage 13/14 and differentiate into the WHPs. Interestingly, the anterior and posterior boundaries of WHP specification coincide with the PS boundaries and WHP specification occurs in register with EPC pairs. This prompted us to study the involvement of Hox genes in defining the boundaries of WHP specification. Based on the published expression patterns, we anticipated to find one of two scenarios: (1) Antp protein expression in PS4 and 5 is sufficient to define the WHP field. Loss of Antp gene function would result in normal levels of Tinman protein expression and, thus, loss of WHPs. (2) Scr defines the anterior and Ubx the posterior boundary. Loss of Scr gene function would expand the WHP field in anterior direction and loss of Ubx gene function in posterior direction. For both scenarios, we would expect to find only Antp protein expression in WHPs while Scr protein should be expressed in the anterior and Ubx protein in the posterior EPCs. Alternatively, Scr or Ubx could act through signaling from the overlying epidermis to the mesodermal EPCs. Consistent with our predicted scenarios, we indeed found only Antp protein expression in WHPs at both investigated stages. However, loss of Antp gene function did not restore Tinman protein expression in WHPs (Figure 4C), which eliminates the first scenario. The second scenario predicts that Scr controls the anterior boundary. However, Scr protein is completely absent from the cardiac mesoderm and therefore also from all EPCs (Lo et al. 2002; Perrin et al. 2004 and this study). Although it is expressed in the epidermis of T1, loss of Scr gene function did not expand the WHP field in anterior direction, which excludes an indirect control. Similarly, Ubx protein was not detected in the EPCs immediately posterior to the WHPs (Zmojdzian et al. 2018, this study). However, loss of Ubx gene function did expand the downregulation of Tinman protein into A1 and A2 leading to 8 WHPs and 8 WHP-like cells. Ubx protein is expressed in the overlaying epidermis at stage 13/14 (Akam and Martinez-Arias 1985) which suggests that Ubx controls the posterior boundary through signaling from the epidermis to the EPCs. Interestingly, only EPCs in A1 and A2 (PS6 and PS7) are converted into additional WHP-like cells in the Ubx null mutant although the Ubx protein expression domain extends to A7 (PS12) in the ectoderm and mesoderm (Akam and Martinez-Arias 1985; Figure 7). Given that Abd-A protein is expressed from the posterior compartment of A2 (PS7) to the anterior compartment of A8 (PS13; Karch et al. 1990), it is very likely that Abd-A also activates Tinman protein expression in the posterior EPCs. However, Ubx and Abd-A protein are expressed in EPCs from A3 to A8 (Zmojdzian et al. 2018) and thus may act cell-autonomously in posterior segments.
Expression pattern of Scr, Antp, Ubx, and abd-A in the epidermis and mesoderm at stage 13/14 based on literature data. At the top, the relationship between segments (S), compartments (a, p), and parasegments (PS) is shown (modified from Akam and Martinez-Arias 1985). Below the location of EPCs in the respective PS is depicted: tin positive EPCs are blue, tin negative EPCs, which become WHPs, are green. The boundaries of WHP specification are marked by dashed lines. Areas of Hox gene expression are depicted as colored bars: Scr—yellow; Antp—green; Ubx—red; abd-A—dark blue. Arabic numerals in front of the bars indicate the literature source: 1—(Martinez-Arias et al. 1987); 2—(Mahaffey and Kaufman 1987); 3—(Riley et al. 1987); 4—(Wirz et al. 1986); 5—(Carroll et al. 1986); 6—(Martinez-Arias 1986); 7—(Akam and Martinez-Arias 1985); 8—(Karch et al. 1990). Cells of the somatic and visceral mesoderm are described to relocate relative to the epidermis from their origin (dashed outline) to their position at stage 13/14 (solid outline). The shift is indicated by an arrow.
This means that also our second scenario did not fully predict the actual situation. Since Scr is not involved in determining the anterior boundary, other Hox genes or homeodomain transcription factors may be responsible for maintaining Tinman protein expression. Like Scr, the anterior three Hox genes of the ANT-C complex, labial, proboscipedia, and Deformed could not be detected in the cardiac mesoderm (Lo et al. 2002; Monier et al. 2007), which makes it very unlikely that they play a role in boundary specification. Another possible candidate, the Hox cofactor and homeodomain transcription factor homothorax, is expressed in the anterior aorta (Perrin et al. 2004). However, the anterior aorta spans all three thoracic segments (T1–T3) which makes homothorax unsuitable to establish a boundary between T1 and T2.
On the other hand, an anterior boundary might not exist. EPCs anterior to T2 (PS4) could simply retain Tinman protein expression from the specification of the cardiac mesoderm (by default Tinman positive) while the EPCs in PS4 and 5 actively loose Tinman protein expression. In his seminal work, Lewis showed that the loss of the entire BX-C transforms all segments posterior to T2 into copies of T2 indicating that T2 is the default state (Lewis 1978), reviewed in Maeda and Karch (2006) and Tomoyasu (2017). A similar observation has been made in the dorsal vessel, where the removal of all Hox function leads to a transformation of the entire heart tube region into anterior aorta identity (Lo et al. 2002; Lovato et al. 2002; Perrin et al. 2004). This means that once the loss of Tinman protein in T2 had been established during evolution, Hox genes had to be recruited to repress this default state in posterior segments. At least for the wing genes, this model seems to hold. Ubx and Abd-A repress wing formation in all abdominal segments. Based on our findings, it is very likely that the same applies to WHP specification since Ubx seems to activate Tinman protein expression in posterior EPCs to repress wing heart formation in the abdomen.
From an evolutionary point of view, it would make perfect sense if wing heart formation was linked to wing formation and both processes were controlled by the same Hox genes. In Drosophila, Antp is the only Hox gene that has the potential to control the differentiation of the forewing due to its expression pattern. However, loss of Antp does not affect forewing morphology (reviewed in Clark-Hachtel and Tomoyasu 2016; Tomoyasu 2017). Consequently, the forewing is considered a Hox-free state which means forewings evolved without the involvement of Hox genes. Our findings suggest the same for the wing hearts. Antp protein is expressed in all WHPs but loss of Antp gene function does not affect their specification. The differentiation of the hindwings into halteres is controlled by Ubx. Loss of Ubx re-establishes the Hox-free ground state and the halteres are transformed into wings. In addition, Ubx/Abd-A repress wing formation in the abdomen (reviewed in Clark-Hachtel and Tomoyasu 2016; Tomoyasu 2017). Our findings support a similar mechanism with respect to the wing hearts. Ubx does not affect WHP specification in T3 but it controls the reduction of the dorsal epidermis and the scutellum in T3. As a result, the WHPs in T3 are relocated into T2 during development. Loss of Ubx re-establishes the notum and scutellum in T3 and WHPs form wing hearts there. Furthermore, Ubx represses WHP specification in A1 and A2, probably also in more posterior segments together with Abd-A. This is supported by the fact that loss of Ubx leads to WHP-like cells in A1 and A2. Taken together, there are striking similarities between Hox-mediated regulation of wing formation and control of wing heart development.
Four-winged flies as a model for the ancestral condition
WHPs are specified in T2 (PS4) and T3 (PS5) even though Drosophila, like all dipterans, only has one pair of functional wings, the forewings, on T2. A plausible explanation for this seemingly unnecessary retention of WHPs in T3 (PS5) may be that, molecularly speaking, dipterans still bear wings on T3 although differentiated into halteres. Studies in Drosophila, Tribolium, and Bombyx have shown, that Ubx is the hindwing selector gene although its function differs in these three insect orders, reviewed e.g., in Tomoyasu (2017). In Drosophila, a main role is the repression of wing genes in T3 halteres, including spalt (sal), vestigal (vg) serum response factor (srf), knirps (kn), and achaete/scute (ac/sc; Akam 1998; Weatherbee et al. 1998; Shashidhara et al. 1999; Mohit et al. 2006; Hersh et al. 2007; Diaz-de-la-Loza et al. 2020). Segments, in which wing formation is completely repressed in the sense that no wing genes are expressed, like in T1 and all abdominal segments, WHPs are completely absent as well. The presence of WHPs in T3 (PS5) prompted us to investigate whether they would be able to form functional wing hearts in an ancestral condition where the wings on T3 are not reduced to halteres. The closest one can get to this ancestral condition, is the use of the famous homeotic transformation that leads to four-winged flies (Lewis 1978, 1998). Even though the result is actually not a T3 with wings but a homeotic copy of T2 (HT2), it allows certain insights into the behavior of WHPs under these conditions. However, for unknown reasons, we were unable to obtain homozygous triple mutants as described by Lewis under our laboratory conditions. Instead, we found an alternative way to generate four-winged flies by bringing the triple mutant over the balancer TM2, Ubx130. About half of these flies exhibit a HT2 with intact notum and scutellum while the other half displays a dorsal cleft along the midline which splits the notum and the scutellum. Due to this separation into left and right side, we termed this phenotype split thorax.
Flies that have been homeotically transformed based on our alternative protocol possess WHPs in T3 (PS4) and T3 (PS5), like wild-type flies, but also WHP-like cells in A1 and in A2. This is very similar to our observation in the embryonic/larval lethal Ubx9.22 null mutants. The weaker phenotype found in four-winged flies (they develop into adulthood) may be explained by the nature of the alleles. In the triple mutant, regulatory elements are affected, which omits expression of Ubx in some but not all parts of the fly, while the null mutant lacks all Ubx function. Completely transformed four-winged flies display four-wing hearts. Functionality of these wing hearts was confirmed by visual observation of wing heart beating in late pupal stages and by the occurrence of successfully cleared wings after eclosion at least in some animals. Live cell imaging revealed that the wing hearts in HT2 originated from WHPs in T3 (PS5) supporting the idea that each set of WHPs in T2 (PS4) and T3 (PS5) are fully capable of forming functional wing hearts.
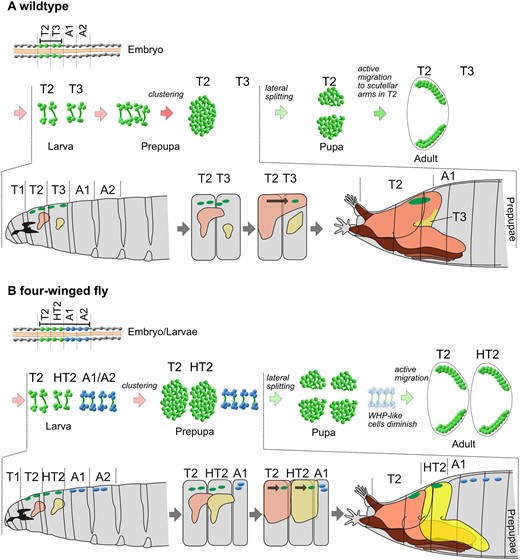
Interestingly, the WHP-like cells in A1 and A2 survive until the late pupal development but do not differentiate into wing hearts in their respective abdominal segment. They also do not relocate into HT2 like one might have expected based on the wild-type development where the WHPs from T3 are relocated into T2 to contribute to wing heart formation. Instead, they remain in their location possibly arrested in development. In addition, four-winged flies with split thoraces displayed varying degrees of completed wing heart formation. Some animals had two wing hearts in HT2 while others had only one wing heart or, in very rare cases, no wing heart at all. These findings combined strongly suggest that the dorsal epidermis of wing-bearing segments provides crucial instructive signals and ultimately determines wing heart formation. Especially, the presence of a scutellum, which later houses the mature wing hearts, may activate and guide the WHPs during their final steps of differentiation. This is already visible in the wild type: The dorsal domains of T2 and T3 in third larval instars are about the same size and the WHPs are located in four clusters directly underneath the epidermis in each of the two segments. However, after the eversion of the imaginal discs at the beginning of pupariation, the entire dorsal thorax consists of cells originating from the T2 wing disc. This is because the wing disc contributes the most cells and spreads the fastest. The epidermal cells from T1 and T3 discs reach the dorsal midline only later during development (Fristrom and Fristrom 1993; Usui and Simpson 2000). This brings all WHPs under the influence of the epidermal domains belonging to T2 and results in the unification of all WHPs in one large cluster. In contrast, in four-winged flies, HT2 also possesses a wing disc from which epidermal cells immediately spread toward the dorsal midline forming a second notum. This spatially separates the WHPs from T2 and HT2 and two clusters are formed (Figure 8). In the split thorax version of the four-winged fly (Figures 5C and 6D), the epidermal cells originating from the wing disc of HT2 spread but the scutellum does not form in the dorsal midline above the WHPs. This leaves the WHPs in HT2 susceptible to the influence of the dorsal domains in T2. Nevertheless, even the bilateral parts of the scutellum in HT2 are able to attract some WHPs and often exhibit functional wing hearts, indicating that the scutella are the driving force behind the subdivision of WHPs by attracting cells in their vicinity. This is further supported by the observation that wild-type WHPs migrate toward the overlap of the pnr-Gal4 and en-Gal4 expressing epidermal domains, which will give rise to the lateral corners of the scutellum in the wild-type (Tögel et al. 2013b).
Model of WHP cluster formation . (A) Wild-type: Eight WHPs (green) are specified in the embryo, located in T2 (PS4) and T3 (PS5). WHPs proliferate during postembryonic stages and are passively relocated into one single cluster during the prepupal stage. This is caused by the eversion of the imaginal discs at the transition from third larval instar to prepupa. The new adult epidermis emerging from the wing disc spreads faster than the one from the neighbouring discs. This leads to a notum of T2 that extends from the head to the abdomen resulting in a temporary discontinuity in segment order. T1 and T3 reach the dorsal midline only later during development. This brings all WHPs under the influence of the T2 notum and they are united in a single cluster. During the pupal stage, the WHPs actively migrate laterally and differentiate into mature wing hearts in the scutellum. (B) Four-winged fly: In addition to the eight WHPs in T2 and T3, eight WHP-like cells (blue) are specified in A1 (PS6) and A2 (PS7). The postembryonic development follows exactly the wild-type pattern except for the clustering during the prepupal stage. The reason for this is the presence of another wing discs in HT2 which spreads at the same speed as the wing disc from T2. This leads to a second large notum in HT2 which keeps the WHPs spatially separated resulting in two clusters. The WHP-like cells in A1 and A2 do not differentiate into wing hearts and disappear. A1, first abdominal segment; A2, second abdominal segment; HT2, homeotically transformed third thoracic segment; T1, first thoracic segment; T2, second thoracic segment; T3, third thoracic segment; WHP, wing heart progenitor. Cartoons of larva and prepupa modified from Hartenstein (1993).
Finally, the observation that wing hearts in four-winged flies are formed from the same pool of WHPs as in the wild-type and that each set of WHPs in T2 and HT2 are fully sufficient to form functional wing hearts strongly suggests that the wild-type WHP anlage in T3 represents the ancestral condition left over from a time when Diptera still had wings on T3 instead of halteres. It further shows that wings, which are exterior dermal derivatives, will only differentiate properly if internal supportive structures and organs co-differentiate. Among these structures and organs are the scutella and the wing hearts (Kiger et al. 2007; Tögel et al. 2008, 2013b; Pass et al. 2015).
Note
Data and some images presented in this article are part of the doctoral thesis of the first author (Tögel 2012).
Data availability
Strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Acknowledgments
The authors thank Martina Biedermann, Mechthild Krabusch, and Lena Dehnen for excellent technical assistance and working as a bachelor student in this project. The authors furthermore thank Rolf Bodmer, Alan Michelson, and Ernesto Sanchez-Herrero for generous gifts of stocks or antibodies. The authors further thank the Developmental Studies Hybridoma Bank and the Bloomington Drosophila Stock Center (NIH P40OD018537) for providing stocks and reagents essential for this work.
Author contributions
Conceptualization: A.P. and M.T.; Validation: A.P. and M.T.; Investigation: M.T.; Writing—original draft: A.P. and M.T.; Writing—review & editing: G.P.; Visualization: A.P., M.T.; Supervision: A.P. and G.P.; Project administration: A.P.; Funding acquisition: A.P.
Funding
This work was supported by grants from the Deutsche Forschungsgemeinschaft DFG to A.P. (SFB 944) and the Austrian Science Fund FWF to G.P. (P19380) and the Osnabrück University (Open Access Publishing Fund).
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
Author notes
Present address: Centre for Neural Circuits and Behaviour, University of Oxford, Tinsley Building, Mansfield Road, Oxford OX1 3SR, UK.



![Cytological location of Ubx and its homeotic mutations. (A) Ubx maps to the cytological band 89D. The homeotic mutations abx, bx3, and pbx are located in the third intron and upstream of the Ubx gene and affect cis regulatory elements [based on FlyBase (Drysdale et al. 2005) and adapted from Maeda and Karch (2006)]. (B) Translocations and inversions induced by irradiation led to a chromosomal breakpoint between band 89D and 89E within the balancer chromosome TM2, Ubx (FlyBase: Drysdale et al. 2005). (C) Flies carrying the triple mutation over the TM2, Ubx balancer display a homeotic transformation of T3 to T2 indicated by the presence of additional wings and a second scutellum. The transformed metathorax (HT2) shows in about 50% of specimens a dorsal cleft named split thorax. The formation of split thoraces is temperature independent. (D) Control and four-winged fly carrying a handC-GFP reporter. In the control (left), the mesothorax is the only large wing bearing segment. At 50 h APF, the wing hearts are visible as bilateral organs in T2 of the pupa. Adult four-winged flies (right) display two large wing bearing segments in the thorax due to the homeotic transformation of T3 to T2 (HT2). The wings are not as perfectly clear as in the control but still show signs of wing maturation. In 50 h APF pupae of completely transformed four-winged flies, two pairs of wing hearts are visible in T2 and HT2. d AH, days after hatching; h APF, hours after puparium formation; T2, second thoracic segment; T3, third thoracic segment; wh, wing heart.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/genetics/220/1/10.1093_genetics_iyab191/1/m_iyab191f5.jpeg?Expires=1716492602&Signature=wZUsC4p3SoedzjWBRHdLXG4A4DCRUVzmtEYwinxog0hE9QZAcLNfAEpI5Y9uL9iOGUYHool~Dkzg7gSF1Jp45c7KhV8PzOr4~Ygz5k6HJPe-rJ1wUXGgjMsA~T1vHg-tt4ZWbr02IYzGoIgTPK7uCUWNeTqegdUYc95v19fJ6T5~TVR9uNXj1vWcE5stJuk3aYZE-IKGVEFOR9UhEpOcD26J0x0pOdUpyPov1hC~Q8FffpbkRsxMfFUUWLj0QT29klgij6WnUtc3UsaoTDkwSAtOGNAQlkMllmO1n0quP~3BF545HjkztdNBjgMvF-oa28lsc3f7YSj0HxwsGbpsHQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)