-
PDF
- Split View
-
Views
-
Cite
Cite
Cynthia M Chai, Wen Chen, Wan-Rong Wong, Heenam Park, Sarah M Cohen, Xuan Wan, Paul W Sternberg, A conserved behavioral role for a nematode interneuron neuropeptide receptor, Genetics, Volume 220, Issue 1, January 2022, iyab198, https://doi.org/10.1093/genetics/iyab198
Close - Share Icon Share
Abstract
Neuropeptides are evolutionarily conserved modulators of many aspects of animal behavior and physiology, and expand the repertoire of processes that can be controlled by a limited number of neurons. Deciphering the neuropeptidergic codes that govern distinct processes requires systematic functional analyses of neuropeptides and their cognate receptors. Even in well-studied model organisms like Caenorhabditis elegans, however, such efforts have been precluded by a lack of mutant reagents. Here, we generated and screened 21 C. elegans neuropeptide G-protein coupled receptor mutants with no pre-existing reagents for the touch-evoked escape response, and implicated six receptors expressed in diverse neuron classes representing multiple circuit levels in this behavior. We further characterized the mutant with the most severe phenotype, frpr-14, which was defective in multiple behavioral paradigms. We leveraged this range of phenotypes to reveal that FRPR-14 modulation of different precommand interneuron classes, AVH and AIB, can drive distinct behavioral subsets, demonstrating cellular context-dependent roles for FRPR-14 signaling. We then show that Caenorhabditis briggsae CBR-FRPR-14 modulates an AVH-like interneuron pair to regulate the same behaviors as C. elegans but to a smaller extent. Our results also suggest that differences in touch-evoked escape circuit architecture between closely related species results from changes in neuropeptide receptor expression pattern, as opposed to ligand–receptor pairing. This study provides insights into the principles utilized by a compact, multiplexed nervous system to generate intraspecific behavioral complexity and interspecific variation.
Introduction
Predation is a major evolutionary selective force that sculpts the form and function of prey circuits. To enhance prey survival chances, escape sensorimotor circuits are designed to execute fast, robust behavioral responses that promote successful capture evasion (Card 2012; Herberholz and Marquart 2012). While foraging, the microscopic Caenorhabditis elegans may encounter predators like nematophagous mites (Karagoz et al. 2007), fungi (Xie et al. 2010; Maguire et al. 2011; Yang et al. 2020), and even other nematode species like Pristionchus pacificus (Bento et al. 2010; Wilecki et al. 2015). Tactile stimulation during these dangerous encounters triggers bidirectional undulatory movements away from the threat that are mediated by C. elegans’ head and body touch-evoked escape circuits (Chalfie and Sulston 1981; Chalfie et al. 1985; Kaplan and Horvitz 1993; Piggott et al. 2011; Pirri and Alkema 2012). The touch-evoked escape response can be modified by different behavioral states (Chao et al. 2004; Schwarz et al. 2011; Cho and Sternberg 2014) and in some cases, neuropeptide signaling has been implicated in driving this state-dependent modulation (Choi et al. 2013; Chen and Chalfie 2014; Nath et al. 2016).
However, a comprehensive understanding of how extrasynaptic neurohormonal networks modulate C. elegans behavior and physiology has been hindered by a lack of mutant reagents. To address this, we used a CRISPR homology-directed repair strategy to generate putative null mutants for the 12 frpr (FMRFamide-like peptide receptor) and 9 npr (neuropeptide receptor) neuropeptide G-protein coupled receptors with no pre-existing mutant reagents (Wang et al. 2018). The npr and frpr families are two of the largest annotated neuropeptide GPCR-encoding gene families in C. elegans and many members have reported behavioral phenotypes (reviewed in Frooninckx et al. 2012). Of the 21 receptor genes selected for knockout in this study, only npr-6 has a previously associated behavioral phenotype (egg-laying defective) identified through RNAi knockdown (Keating et al. 2003).
Materials and methods
Animal maintenance and strains
Animals were cultivated at 20°C on standard nematode growth media (NGM) plates seeded with Escherichia coli OP50 cultured in Luria-Bertani broth. The following strains were used in this study:
N2 (Bristol)
AF16 (India)
PS8429: frpr-14(sy1301)
PS8430: frpr-17(sy1302)
PS8487: frpr-8(sy1363)
PS8400: frpr-7(sy1296)
PS8484: npr-31(sy1360)
PS8450: npr-27(sy1315)
PS8490: frpr-16(sy1366)
PS8432: frpr-19(sy1304)
PS8488: frpr-2(sy1364)
PS8398: frpr-9(sy1294)
PS8315: npr-29(sy1270)
PS8444: npr-21(sy1309)
PS8442: npr-26(sy1307)
PS8330: frpr-5(sy1274)
PS8426: frpr-13(sy1298)
PS8317: npr-33(sy1272)
PS8396: frpr-1(sy1292)
PS8177: npr-23(sy1203)
PS8334: frpr-11(sy1278)
PS8938: frpr-12(sy1581)
PS8902: npr-6(sy1571)
PS7379: flp-3(ok3265)
PS9050: flp-4(sy1606)
VC2324: flp-6(ok3056)
PS9098: flp-3(ok3265); frpr-14(sy1301)
PS9056: flp-4(sy1612); frpr-14(sy1301)
PS9099: flp-6(ok3056); frpr-14(sy1301)
PS9138: syEx1857 [Pfrpr-1::GFP; Pnmr-1::mCherry-H2B, Pofm-1::RFP]
PS9139: syEx1858 [Pfrpr-5::GFP; Pnmr-1::mCherry-H2B, Pofm-1::RFP]
PS9140: syEx1859 [Pfrpr-12::GFP; Pnmr-1::mCherry-H2B, Pofm-1::RFP]
PS9141: syEx1860 [Pnpr-29::GFP; Pnmr-1::mCherry-H2B, Pofm-1::RFP]
PS9142: syEx1861 [Pnpr-31::GFP; Pnmr-1::mCherry-H2B, Pofm-1::RFP]
PS9146: syEx1862 [Pflp-3::GFP; Pmec-4::mCherry-H2B, Pofm-1::RFP]
PS8948: syEx1789 [Pfrpr-14::frpr-14 gDNA::SL2::GFP, Phlh-34::mCherry-H2B, Pofm-1::RFP]; sy1301
PS8927: syEx1785 [Pfrpr-14::frpr-14 gDNA::SL2::GFP, Pnpr-9::mCherry-H2B, Pofm-1::RFP]; sy1301
PS8957: syEx1795 [Phlh-34::frpr-14 cDNA::SL2::GFP, Pofm-1::RFP]; sy1301
PS8946: syEx1787 [Pnpr-9::frpr-14 cDNA::SL2::GFP, Pofm-1::RFP]; sy1301
PS8984: syEx1803 [Phlh-34::Cbr-frpr-14 cDNA::SL2::GFP, Pofm-1::RFP]; sy1301
PS9003: syEx1808 [Phlh-34::Bma-frpr-14 cDNA::SL2::GFP, Pofm-1::RFP]; sy1301
PS9103: syEx1841 [Phlh-34::Ovo-frpr-14 cDNA::SL2::GFP, Pofm-1::RFP]; sy1301
PS9001: Cbr-frpr-14(sy1601)
PS9000: Cbr-frpr-14(sy1602)
PS8977: syEx1796 [PCbr-frpr-14::Cbr-frpr-14 gDNA::SL2::GFP]
CB1490: him-5(e1490)
CB169: unc-31(e169)
Molecular biology and transgenesis
frpr-14 cDNA was PCR amplified from N2 cDNA library. Gene promoters and frpr-14 genomic DNA were PCR amplified from N2 genomic DNA library. Cbr-frpr-14 genomic DNA was PCR amplified from AF16 genomic DNA library. Promoters for generating transcriptional reporters are hlh-34 (2.6 kb), npr-9 (2.0 kb), frpr-1 (2.5 kb), frpr-5 (2.5 kb), frpr-12 (1.1 kb), npr-29 (2.5 kb), npr-31 (3.0 kb), nmr-1 (4.7 kb), mec-4 (250 bp), and flp-3 (1.5 kb). The entire C. elegans frpr-14 genomic locus including 600 bp of regulatory region upstream of the start codon was used to generate the frpr-14 translational GFP fusion. The entire C. briggsae Cbr-frpr-14 genomic locus including 570 bp of regulatory region upstream of the start codon was used to generate the Cbr-frpr-14 translational GFP fusion. Cbr-frpr-14 (CBG19545), Bma-frpr-14 (Bm4879a), and Ovo-frpr-14 (OVOC11602) cDNA constructs were synthesized by Integrated Data Technologies gBlocks. Constructs were inserted into either pSM GFP or pSM mCherry-H2B vector backbone using HiFi Assembly or restriction enzyme cloning.
Transgenic strains were generated by microinjection of plasmids with coinjection markers into adult worms. cDNA plasmids were injected at 50 ng/µl. GFP and mCherry reporter constructs were injected at 25–50 ng/µl. Coinjection marker Pofm-1::RFP was injected at 40 ng/µl. One kilobase of DNA ladder was used as a filler to bring final DNA concentration to 200 ng/µl.
CRISPR mutagenesis
CRISPR mutagenesis in C. elegans was performed as previously described (Wang et al. 2018). CRISPR mutagenesis in C. briggsae was performed as previously described (Cohen and Sternberg 2019). Briefly, a 43-bp universal STOP-IN cassette was inserted near the target gene’s 5′ end to disrupt translation. Independent F2 homozygous animals were isolated using PCR detection and confirmed with Sanger sequencing.
Posterior body gentle touch-evoked escape response assay
Ten L4 stage worms were picked to seeded NGM plates and allowed to lay eggs for 2 h before being removed. Plates were incubated at 20°C until worms reached the young adult stage. On each plate, ten worms were scored individually for the posterior body touch-evoked escape response. Using an eyelash pick, each worm was gently touched on the anterior body region immediately posterior to the pharynx to trigger a reversal. Immediately after the reversal motor program terminated, worms were touched on the posterior body region. Whether or not the worm responded to this posterior touch by moving forward was recorded. This procedure was repeated 10 times per animal with a time interval of 5 s in between trials. The response rate for each worm is the percentage of positive responses (forward movements) out of 10 trials. The average response per batch is the average of response rates for the ten worms assayed per plate. At least three batches were assayed per strain. All assays were conducted at 22°C.
Two independent F1 transgenic lines were assayed for each touch-evoked escape rescue experiment. In all touch-evoked escape rescue experiments, both independent F1 transgenic lines rescued the frpr-14(−) mutant’s defect to similar extents (not statistically significant from each other). We then selected the F1 transgenic line with the highest array transmission rate for further investigation using the motility assay described below.
Motility assay
L4 stage animals were picked around 16 h before experiments and grown at 20°C overnight. On the day of experiments, experimental plates were seeded with a saturation-phase culture of E. coli OP50. Plates were left to dry at room temperature for 1–2 h. Four to six young adults were then picked onto each experimental plate and left to acclimatize for at least 30 min at room temperature. Each plate was then video recorded for 4 min at 7.5 frames/s using the MBF Bioscience WormLab setup. The camera field of view was centered on each food patch. All recordings were conducted within 30–60 min after worms were transferred to experimental plates. Videos were analyzed using WormLab software with the following mobility idle time analysis settings—speed threshold: 1.5 µm/s, averaging window: five frames, minimum duration: five frames. As animals sometimes move out of the field of view during video recording, each animal’s total mobility idle time was normalized by total track duration to obtain the percentage of mobility idle time. All assays were conducted at 22°C.
Male mating assays
L4 stage males and L4 stage unc-31 hermaphrodites were picked to separate seeded NGM plates and grown at 20°C overnight. NGM plates were spotted at the center with 10 µl of concentrated E. coli OP50 and dried overnight at room temperature. On the day of experiments, 15 adult unc-31 hermaphrodites were picked onto the bacteria lawn of each mating plate. Three adult males were then placed onto the bacteria lawn with the hermaphrodites and behavior was video recorded for 4 min. The mating behaviors of each male were manually scored throughout this duration using the following criteria:
Touch response assay
Touch not leading to mating response: ventral side of male tail makes touch contact with hermaphrodite but male does not initiate backing.
Touch leading to mating response: ventral side of male tail makes touch contact with hermaphrodite and male initiates backing.
Vulva location assay
Successful vulva location: male stops for >1 s at vulva.
Vulva pass: male does not stop at vulva.
Hesitation: male pauses at vulva for <1 s but then moves away.
All assays were conducted at 22°C.
Microscopy
Animals were immobilized with 100 mM sodium azide on 2% agarose pads. Slides were imaged under a ZEISS Axio Imager.Z2 microscope attached to a Axiocam 506 mono microscope camera. Images were processed using ZenPro software.
Statistical analysis
For touch-evoked escape assays, unpaired Welch’s T-test or ANOVA followed by Tukey HSD post hoc analysis were used to determine statistically significant differences between groups. For motility assays, Kruskal–Wallis test followed by Dunn’s post hoc with Holm–Bonferroni correction was used to determine statistically significant differences between groups. For male-mating assays, Wilcoxon rank sum test was used to determine statistically significant differences between groups. All P-values reported in the figure legends are corrected P-values. Statistical analyses were performed in R. Plots were generated in Python.
Results and discussion
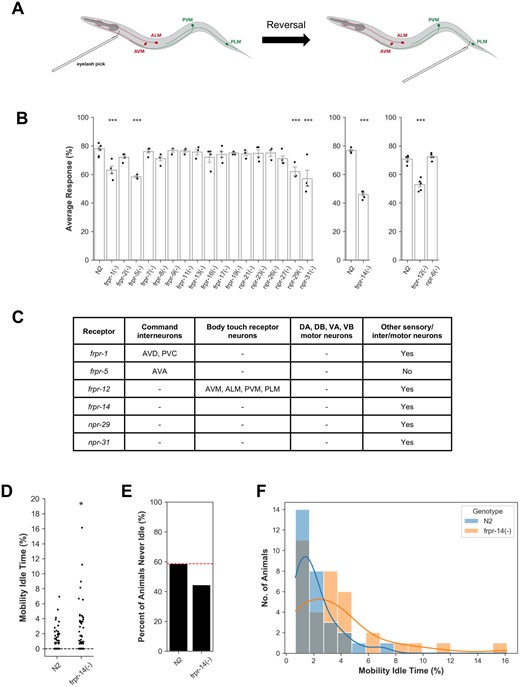
To generate neuropeptide GPCR mutants, a triple stop cassette was inserted in-frame near the 5′ end of each predicted receptor-encoding gene sequence. All 21 neuropeptide receptor mutants generated here did not exhibit observable abnormalities in developmental growth rate. We behaviorally screened this set of mutants using the posterior body gentle touch-evoked escape response assay. In this assay, an eyelash pick was used to gently stroke the animal’s anterior body region immediately behind the pharynx to elicit a reversal (Figure 1A). At the end of the reversal sequence, another gentle touch stimulus is delivered to the tail region within the PLM touch receptor neurons’ sensory field, and whether or not a forward movement occurs is recorded (Figure 1A). This procedure was repeated ten consecutive times for each animal and the percentage of forward movement responses per animal is calculated. The response rates of 10 animals were then averaged to obtain each batch average response. Our screen revealed touch-evoked escape defective phenotypes for six receptor mutants—frpr-1, frpr-5, frpr-12, frpr-14, npr-29, and npr-31 (Figure 1B). We next generated transcriptional GFP reporters for these receptors and analyzed the extent of expression pattern overlap with the body gentle touch-evoked escape circuit consisting of the ALM, AVM, PVM, PLM touch sensors, the AVA, AVB, AVD, PVC command interneurons, and motor neurons (Chalfie and Sulston 1981; Chalfie et al. 1985; Chatzigeorgiou et al. 2010). While frpr-5 and frpr-12 expression is largely restricted to canonical circuit elements such as premotor command interneurons and body touch receptor neurons, the other four receptors are expressed in diverse neuron classes including chemosensory neurons and noncommand interneurons (Figure 1C). Taken together, these observations indicate that the neural circuitry underlying the body touch-evoked escape response is more distributed than previously thought and is modulated at multiple circuit levels by neuropeptide signaling.
Multilevel modulation of the body touch-evoked escape circuit by neuropeptide signaling. (A) Schematic of posterior body gentle touch-evoked escape assay. Drawing not to scale. (B) Touch-evoked escape response assay screen of neuropeptide GPCR putative null mutants. Error bars represent standard error of the mean. N = 3–8 batches. One-way ANOVA followed by Dunnett’s post hoc compared with N2. Unpaired Welch’s t-test for N2—frpr-14(−) pairwise comparison. ***P < 0.001. frpr-1(−) (P = 1.7e−04), frpr-5(−) (P = 5.7e−06), frpr-12(−) (P = 4.5e−08), frpr-14(−) (P = 1.0e−08), npr-29(−) (P = 3.0e−04), npr-31(−) (P = 5.9e−08). (C) Summary of touch-evoked escape defective receptor expression patterns. Data derived from frpr-1, frpr-5, frpr-12, npr-29, npr-31 transcriptional GFP reporter strains coexpressing a Pnmr-1::mCherry-H2B command interneuron marker (Zheng et al. 1999), and frpr-14 translational GFP reporter strains coexpressing either a Phlh-34::mCherry-H2B AVH-specific marker (Cook et al. 2021) or a Pnpr-9::mCherry-H2B AIB-specific marker (Bendena et al. 2008) (see Supplementary Fig S2, A and B for images). Presence or absence of AVB interneurons determined by soma position relative to AVD (as marked by Pnmr-1::mCherry-H2B) and process morphology. (D) Percentage of mobility idle time throughout recording duration. N = 63–70 animals. Wilcoxon rank sum test. *P < 0.05. P = 2.622e−02. (E) Percent of animals in (D) that were never idle (i.e., Mobility Idle Time = 0). (F) Distribution of nonzero mobility idle times in (D). Colored lines represent kernel density estimation. N = 29–35 animals.
As frpr-14 mutants had the most severe touch-evoked escape response defect compared with control animals, we chose to further characterize the neurobehavioral role of this receptor. During food patch exploration in search of mating partners, C. elegans males use specialized tail ray structures to detect hermaphrodite-specific structural and chemical features leading to initiation and execution of the male mating behavior sequence (Liu and Sternberg 1995; Barr and Sternberg 1999). Upon touch contact with and successful recognition of the hermaphrodite, the male uses its tail to travel along the hermaphrodite’s body length until it locates the vulva and copulates (Liu and Sternberg 1995). We found that frpr-14 mutant males are deficient at initiating the mating sequence upon touch contact with the hermaphrodite (Supplementary Figure S1, A and B) and vulva location (Supplementary Figure S1, C and D) demonstrating a sexually dimorphic role for FRPR-14 in male mating.
During routine animal maintenance, we observed that frpr-14 mutants exhibit prolonged bouts of locomotion quiescence compared with wild-type N2 animals. Using an automated freely moving behavior recording and analysis setup, we quantified this spontaneous motility defect as the percentage of recording time the animal did not exceed a specified bidirectional movement distance threshold (henceforth called “mobility idle time”). Under these conditions, frpr-14 mutants show a significantly greater mobility idle time relative to controls (Figure 1, D–F). We also note that while frpr-14 mutants are able to spontaneously execute bidirectional movement, they are slow and uncoordinated when so doing.
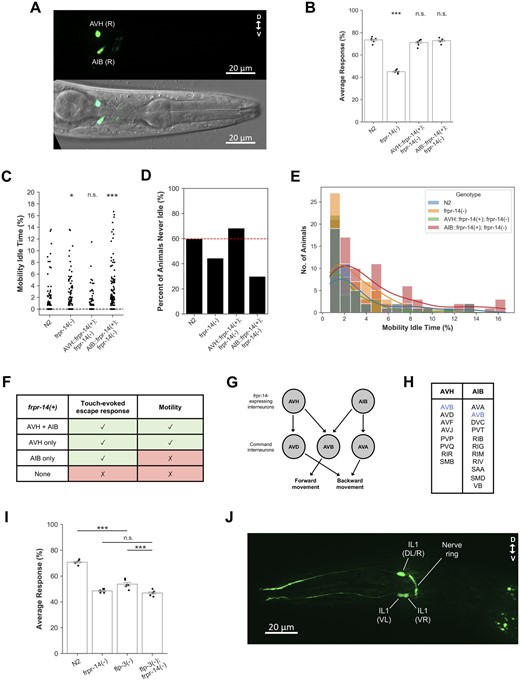
Based on colocalization analysis using AVH- and AIB-specific markers, respectively, we determined that a frpr-14 translational GFP reporter was expressed in the AVH and AIB interneuron classes (Figure 2A;Supplementary Figure S2, A and B) (Bendena et al. 2008; Cook et al. 2021). Neither neuron class has a reported role in the body touch-evoked escape response. To determine receptor site of action, we expressed functional copies of frpr-14 in a neuron class-specific manner in frpr-14 mutants and assayed transgenic animals using both the touch-evoked escape response and motility assays. We found that either AVH- or AIB-specific expression was sufficient to fully reinstate the mutant’s touch-evoked escape response (Figure 2B), while only AVH-specific expression rescued the mutant’s motility defect back to control levels (Figure 2, C–E). This site-of-action analysis revealed overlapping roles for frpr-14-expressing interneurons with AVH exerting more comprehensive control over the behavioral repertoire examined in this study compared with AIB (Figure 2F). An examination of the synaptic connections between frpr-14-expressing interneurons and the premotor command interneurons provides some clues as to how this cellular context-dependent phenotypic specificity is achieved. AVH and AIB synaptic inputs converge only on AVB, which drives forward movement together with PVC (Figure 2G) (Chalfie et al. 1985; White et al. 1986). Other than AVB, both frpr-14-expressing interneurons do not share any significant downstream synaptic targets (Figure 2H) (White et al. 1986). Furthermore, AVH and AIB do not form any synapses with each other (White et al. 1986). In the absence of AVH modulation, FRPR-14-mediated AIB activity can act as an alternate driver of the escape response but not spontaneous motility perhaps due to the lack of necessary connections with an AVH-specific motility circuit module. From an ethological perspective, the functional redundancy of AVH and AIB in the escape response but not spontaneous motility makes sense as successful timely escapes from predators is critical for survival while disrupted locomotion patterns during foraging for resources does not pose any imminent danger. An additional network of neuropeptide-mediated extrasynaptic connections might also exist between the frpr-14-expressing interneurons and the command interneurons, potentially modulating the cellular responses of this integrative processing layer.
FRPR-14 modulates precommand interneuron function during touch-evoked escape response and motility behaviors. (A) Representative image of frpr-14 translational GFP reporter expression in L4 stage animal. Top—GFP only. Bottom—DIC and GFP merge. (B) Touch-evoked escape response results for AVH and AIB-specific frpr-14 cDNA expression in frpr-14(−) background. Error bars represent standard error of the mean. N = 7 batches. One-way ANOVA followed by Tukey’s HSD post hoc. Only statistical comparisons to N2 shown. ***P < 0.001, n.s., no significance. N2—frpr-14(−) (P = 2.109e−14), N2—AVH::frpr-14(+); frpr-14(−) (P = 2.264e−01), N2—AIB::frpr-14(+); frpr-14(−) (P = 9.360e−01). (C) Percentage of mobility idle time throughout recording duration. N = 117–120 animals. Kruskal–Wallis test followed by Dunn’s post hoc (Holm–Bonferroni correction). Only statistical comparisons to N2 shown. *P < 0.05; ***P < 0.001; n.s., no significance. N2—frpr-14(−) (P = 3.924e−02), N2—AVH::frpr-14(+); frpr-14(−) (P = 1.108e−01), N2—AIB::frpr-14(+); frpr-14(−) (P = 1.123e−06). (D) Percent of animals in (C) that were never idle (i.e., Mobility Idle Time = 0). (E) Distribution of nonzero mobility idle times in (C). Colored lines represent kernel density estimation. N = 38–84 animals. (F) Summary of frpr-14 site-of-action analysis. (G) Schematic showing synaptic connections between frpr-14-expressing interneurons and command interneurons (White et al. 1986). Arrows represent chemical synapses. Connections are unweighted. (H) Interneuron and motorneuron classes postsynaptic to AVH and AIB (White et al. 1986). Only connections with more than one chemical and/or electrical synapse included. (I) flp-3(−); frpr-14(−) genetic epistasis analysis for touch-evoked escape response. Error bars represent standard error of the mean. N = 6 batches. One-way ANOVA followed by Tukey’s HSD post hoc. ***P < 0.001; n.s., no significance. N2—flp-3(−) (P = 1.942e−10), flp-3(−)—flp-3(−); frpr-14(−) (P = 2.279e−04), frpr-14(−)—flp-3(−); frpr-14(−) (P = 5.904e−01). (J) Representative image of flp-3 transcriptional GFP reporter expression in L4 stage animal. Expression in IL1(L/R) is highly variable, and when present, very weak.
The C. elegans frpr gene family is related to the Drosophila FMRFamide receptor FR, and members of the frpr family are predicted to encode cognate receptors for FMRFamide-like peptides encoded by the flp gene family (Meeusen et al. 2002; Hobert 2013). To identify potential frpr-14 interactors, we first screened 26 flp mutants by visual inspection for a similar decreased motility phenotype as frpr-14 mutants (Supplementary Table S1). Three candidates, flp-3, flp-4, and flp-6, were then challenged with the touch-evoked escape response assay and we found that all three are defective relative to control animals. Double mutant epistasis analysis revealed that flp-4(−); frpr-14(−) and flp-6(−); frpr-14(−) animals had significantly enhanced touch-evoked response defects compared with the single mutants (Supplementary Figure S3, A and B) while flp-3(−); frpr-14(−) animals phenocopied the frpr-14 mutant’s phenotype (Figure 2I). This suggests that flp-3 is in the same genetic pathway as frpr-14 and likely upstream based on the nature of their gene products. In addition to other neuron classes, flp-3 is expressed in the IL1 head mechanosensory neurons (Figure 2J) (Kim and Li 2004), which regulate the head-withdrawal reflex in response to gentle touches to the side of the animal’s head via the RMD motor neurons (Hart et al. 1995). Laser microbeam ablation of IL1 also decreases spontaneous head oscillations during foraging (Hart et al. 1995). Interestingly, IL1 does not make any synaptic connections with the frpr-14-expressing AVH and AIB interneurons (White et al. 1986). Thus, a pathway involving flp-3 and frpr-14 might couple anatomically distinct touch-evoked escape and motility circuits to coordinate appropriate whole-animal behavioral responses. Further experiments will need to be performed to determine if FLP-3 and FRPR-14 directly interact at the protein level.
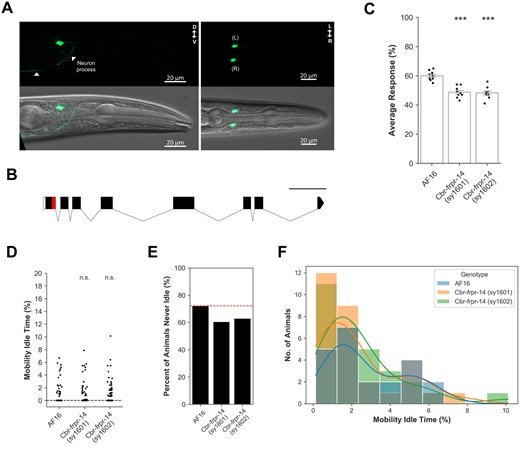
Caenorhabditis briggsae is a closely related free-living species that diverged from C. elegans at least 18 million years ago (Stein et al. 2003; Cutter 2008). Although comparative developmental studies between both species have described conserved and divergent regulatory mechanisms, much less is known about how their neurobehavioral programs are related (Rudel and Kimble 2001; Haag et al. 2002; Wang and Chamberlin 2002; Kirouac and Sternberg 2003; Baird et al. 2005; Nayak et al. 2005; Inoue et al. 2007). We used the C. briggsae AF16 tropical strain as our reference strain (Stein et al. 2003). We wondered if C. briggsae’s receptor ortholog Cbr-frpr-14 regulates the same behaviors as in C. elegans and whether Cbr-frpr-14 modulates the function of similar neuron types (Supplementary Figure S4). Cbr-frpr-14 is expressed in a single bilaterally symmetric pair of interneurons with cell bodies located in the same relative anatomical positions as the AVH interneurons in C. elegans (Figure 3A) (White et al. 1986). The processes of these C. briggsae AVH-like interneurons also enter the nerve ring before extending into the ventral nerve cord (Figure 3A) (White et al. 1986). To determine the physiological relevance of this receptor in C. briggsae, we again applied a CRISPR homology-directed repair strategy to generate Cbr-frpr-14 putative null mutants (Figure 3B) (Cohen and Sternberg 2019). We then assayed these mutants using the touch-evoked escape response and motility assays under the same experimental conditions as C. elegans. Both Cbr-frpr-14 mutant strains had a significantly decreased touch-evoked escape response compared with the C. briggsae AF16 wild-type strain, and this difference was more subtle compared with C. elegans (Figure 3C). Although overall Cbr-frpr-14 mutant mobility idle time was not significantly different from control animals, the percentage of Cbr-frpr-14 mutants that were never idle was consistently lower than the AF16 wild-type control (Figure 3, D–F). Thus, FRPR-14 receptor orthologs modulate the function of a homologous interneuron class to regulate the same behaviors in two Caenorhabditis sibling species to different extents. Assuming that all relevant Cbr-frpr-14 cis-regulatory elements are present in our translational reporter construct, Cbr-frpr-14 expression in only an AVH-like pair indicates that there is no alternative CBR-FRPR-14-mediated neural pathway to drive the C. briggase touch-evoked escape response in the absence of the AVH-like neurons’ receptor-mediated functionality. We speculate that either this circuit degeneracy has been selected against in C. briggsae’s natural environment or other receptor–neuron pairing(s) play this role in C. briggsae.
Cbr-frpr-14 regulates the same behaviors in C. briggsae via modulation of an AVH-like interneuron pair. (A) Representative image of Cbr-frpr-14 translational GFP reporter expression in young adult stage animal. Top—GFP only. Bottom—DIC and GFP merge. (B) Schematic depicting stop cassette insertion site (red) near the end of Cbr-frpr-14’s first exon. Scale bar represents 500 bp. (C) Touch-evoked escape response assay for Cbr-frpr-14 null mutants. Error bars represent standard error of the mean. N = 9 batches. One-way ANOVA followed by Dunnett’s post hoc compared with AF16. ***P < 0.001. Cbr-frpr-14 (sy1601) (P = 2.8e−06), Cbr-frpr-14 (sy1602) (P = 1.6e−06). (D) Percentage of mobility idle time throughout recording duration. N = 76–81 animals. Kruskal–Wallis test followed by Dunn’s post hoc (Holm–Bonferroni correction). Only statistical comparisons to N2 shown. n.s., no significance. AF16—Cbr-frpr-14 (sy1601) (P = 7.273e−01), AF16—Cbr-frpr-14 (sy1602) (P = 6.402e−01). (E) Percent of animals in (D) that were never idle (i.e., Mobility Idle Time = 0). (F) Distribution of nonzero mobility idle times in (E). Colored lines represent kernel density estimation. N = 22–30 animals.
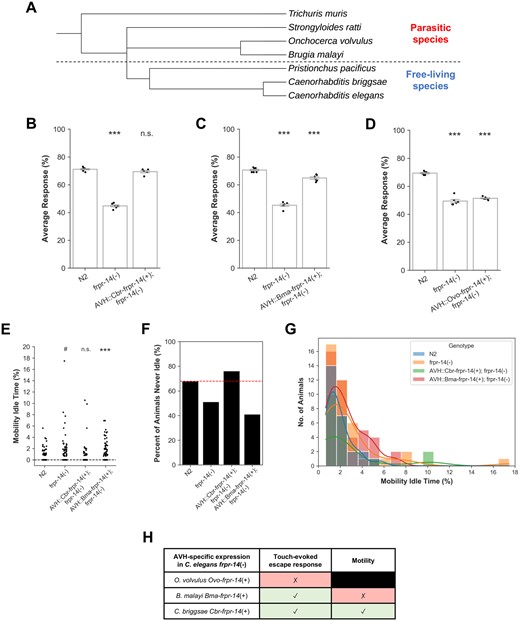
Despite the high conservation of the FLP peptide repertoire between Caenorhabditis species (Li and Kim 2014), whether orthologous ligand–receptor relationships are also conserved has never been experimentally probed in vivo. We found that AVH-specific Cbr-frpr-14 cDNA expression in the C. elegans frpr-14 mutant was sufficient to fully rescue both touch-evoked escape and motility defects suggesting full conservation of this ligand–receptor interaction between both Caenorhabditis species (Figure 4, B and E–G). Electron microscopy-based reconstruction of the P. pacificus sensory nervous system indicates that P. pacificus and C. elegans, which diverged ∼100 million years ago, share similar overall synaptic connectivity structures (Hong et al. 2019). How then might numerically constrained nervous systems reconfigure sensorimotor information flow to generate species-specific behavioral variation during evolutionary adaptation? Changes in neuropeptide ligand/receptor coding sequences that affect their protein interactions and changes in cis/trans-regulatory elements that determine ligand/receptor spatial expression patterns represent two alternative circuit “rewiring” strategies impinging upon extrasynaptic neuropeptide signaling networks. Our results suggest that, in the context of the touch-evoked escape circuit, the latter strategy is employed. Cis/trans-regulatory evolution has also been previously reported to underlie phenotypic divergence between sibling species of both Caenorhabditis and Drosophila (Sucena and Stern 2000; Wang and Chamberlin 2002; Ortiz et al. 2006; Nagy et al. 2018; Prabh et al. 2018).
FRPR-14 ortholog-mediated C. elegans AVH behavioral output range decreases with evolutionary distance. (A) Phylogenetic tree showing selected free-living and parasitic species of the nematode phylum. Touch-evoked escape response assay for cross-species AVH-specific Cbr- or Bma- or Ovo-frpr-14 cDNA expression in C. elegans frpr-14(−) background. Error bars represent standard error of the mean. N = 5–6 batches. One-way ANOVA followed by Tukey’s HSD post hoc. Only statistical comparisons to N2 shown. ***P < 0.001; n.s., no significance. (B) Caenorhabditis briggsae Cbr-frpr-14 cDNA. N2—frpr-14(−) (P = 9.155e−13), N2—AVH::Cbr-frpr-14(+); frpr-14(−) (P = 2.813e−01). (C) Brugia malayi Bma-frpr-14 cDNA. N2—frpr-14(−) (P = 5.531e−12), N2—AVH::Bma-frpr-14(+); frpr-14(−) (P = 7.808e−04). (D) Onchocerca volvulus Ovo-frpr-14 cDNA. N2—frpr-14(−) (P = 5.445e−11), N2—AVH::Ovo-frpr-14(+); frpr-14(−) (P = 2.458e−10). (E) Percentage of mobility idle time throughout recording duration. N = 73–84 animals. Kruskal–Wallis test followed by Dunn’s post hoc (Holm–Bonferroni correction). Only statistical comparisons to N2 shown. #P < 0.1; **P < 0.01; n.s., no significance. N2—frpr-14(−) (P = 7.256e−02), N2—AVH::Cbr-frpr-14(+); frpr-14(−) (P = 3.186e−01), N2—AVH::Bma-frpr-14(+); frpr-14(−) (P = 1.1556e−03). (F) Percent of animals in (E) that were never idle (i.e., Mobility Idle Time = 0). (G) Distribution of nonzero mobility idle times (F). Colored lines represent kernel density estimation. N = 20–43 animals. (H) Summary of C. elegans frpr-14(−) AVH-specific frpr-14 ortholog cDNA expression analysis.
As a subset of C. elegans flp genes, including flp-3, have sequelogs in parasitic species (McCoy et al. 2014), we further extended our cross-species receptor expression analysis to frpr-14 orthologs of the filarial parasites Brugia malayi and Onchocerca volvulus (Figure 4A). Surprisingly, we observed almost full (∼77%) rescue of the mutant’s touch-evoked escape response defect by AVH-specific Bma-frpr-14 expression but a complete absence of a motility defect rescue (Figure 4, C and E–G). The simplest explanation for this result is that differences between C. elegans and B. malayi orthologous receptor binding site residues lead to decreased C. elegans endogenous neuropeptide ligand efficacy (partial agonism) and weaker activation of BMA-FRPR-14 (Weis and Kobilka 2018). This difference could have shifted AVH neural activity in excess of some threshold to partially trigger the escape response but was insufficient to affect the mutant’s motility defect. AVH-specific expression of the most distantly related Ovo-frpr-14 cDNA did not rescue the C. elegans frpr-14 mutant’s touch-evoked response defect (Figure 4D). Although this study does not further pursue the mechanisms underlying the functional differences between these receptor orthologs (Figure 4H), we propose this experimental paradigm as a tractable in vivo system for future studies on how related receptor sequences expressed in the same cellular context can produce specific behavioral outputs.
Through systematic behavioral screening of neuropeptide receptor mutants followed by expression pattern analysis of candidate receptors, we have implicated new neuron classes in the circuitry controlling the C. elegans body touch-evoked escape response. Our approach and findings underscore the value of functional genetic screens in the discovery of novel circuit elements. We further show that the receptor mutant with the most severe phenotype, frpr-14, also has a spontaneous motility defect and that FRPR-14 signaling via different interneuron classes can drive distinct behavioral subsets. Future studies will elucidate whether and how this cellular context-dependent phenotypic specificity arises from the interplay between receptor site of action and the animal’s hardwired neural architecture.
By applying precision genome editing to perturb Cbr-frpr-14 function in C. briggsae, we directly show that orthologous interneuron neuropeptide receptors regulate the same behaviors in two closely related Caenorhabditis species but to different extents. Our results also suggest that changes in neuropeptide receptor expression, but not ligand–receptor interactions, underlie species-specific differences in touch-evoked escape circuit architecture. Synapse formation, maintenance, and information transmission are metabolically costly (Laughlin 2001; Laughlin and Sejnowski 2003). Extrasynaptic neuropeptide signaling helps to minimize this metabolic load by “wirelessly” expanding short and long-range neural connectivity and by increasing the modularity of the hardwired connectome. By examining the function and site of action of an interneuron neuropeptide receptor along two dimensions, behavior and species, we have shed light on the principles used by a compact brain to generate intraspecific behavioral complexity and interspecific variation.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplementary material is available at GENETICS online.
Acknowledgments
Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). WormBase provided key molecular and genetic information. Some strains were provided by the lab of Dr Shohei Mitani as part of the National Bioresource Project. We thank Shu Fay Ung for generating the schematic in Figure 1A.
Conceptualization—C.M.C. and P.W.S. Experimental design and data analysis—C.M.C., W.C., W.W., and X.W. Molecular cloning and transgenesis—C.M.C. Caenorhabditis elegans CRISPR mutagenesis—H.P. Caenorhabditis briggsae CRISPR mutagenesis—S.M.C. Touch-evoked escape response assays—W.C. Motility assays—W.W. Male mating assays—X.W. Microscopy and neuron identification—C.M.C. C.M.C. wrote the paper with input from all coauthors. Funding acquisition—P.W.S.
Funding
This work was funded by NIH grants R240D023041 and R01NS113119 to P.W.S.
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
Author notes
Cynthia M. Chai, Wen Chen and Wan-Rong Wong contributed equally to this work.