-
PDF
- Split View
-
Views
-
Cite
Cite
Dafang Wang, Chuanhe Yu, Jianbo Zhang, Thomas Peterson, Excision and reinsertion of Ac macrotransposons in maize, Genetics, Volume 221, Issue 4, August 2022, iyac067, https://doi.org/10.1093/genetics/iyac067
Close - Share Icon Share
Abstract
Eukaryotic Macrotransposons (MTns) can be formed by 2 nearby elements flanking a segment of host DNA. The maize Ac transposon can form Ac::MTns, but little is known about Ac::MTn transposition activities. Here, we studied 3 Ac::MTns at the maize p1 locus, each of which is composed of a segment of maize p1 genomic DNA (up to 15 kb) bounded by a fractured Ac element (fAc, 2039 bp), and a full-length Ac element in direct orientation. The resulting Ac::MTns are of 16, 16.5, and 22 kb total length. From these 3 Ac::MTns, we identified 10 independent cases of macrotransposition, and observed similar features of transposition between Ac::MTn and standard Ac/Ds, including characteristic excision footprints and insertion target site duplications. Nine out of the 10 Ac::MTn reinsertion targets were genetically linked to the donor sites, another similarity with Ac/Ds standard transposition. We also identified a MTn-like structure in the maize B73 reference genome and 5 NAM founder lines. The MTn in diverse lines is flanked by target site duplications, confirming the historic occurrence of MTn transposition during genome evolution. Our results show that Ac::MTns are capable of mobilizing segments of DNA long enough to include a typical full-length plant gene and in theory could erode gene colinearity in syntenic regions during plant genome evolution.
Introduction
Ac (Activator) and Ds (Dissociation) were the first transposable elements discovered and described by Barbara McClintock in the 1940s (McClintock 1948, 1950, 1951). As Class II transposons in the hAT superfamily, Ac/Ds elements utilize a “cut and paste” mechanism of transposition. The autonomous Ac element is 4,565 bp in length, contains 11-bp imperfect terminal inverted repeats (TIRs), and encodes a single transposase (TPase) gene. Fractured Ac (fAc) and nonautonomous Ds elements commonly lack a complete TPase coding sequence; however, they retain one or both functional transposon termini, respectively, and are capable of transposition in the presence of Ac (Kunze et al. 1987; Kunze and Starlinger 1989; Kunze and Weil 2002).
Standard Ac transposition only involves the termini of a single element. During transposition, Ac/Ds elements produce excision footprints at donor sequences, often characterized by local nucleotide changes; i.e. one to several basepairs deletion, transition or transversion (Rinehart et al. 1997). Ac/Ds insertion creates 8-bp target site duplications (TSDs) that flank the newly inserted element (Kunze and Weil 2002). Ac/Ds transposition also shows a high tendency for insertion into local, genetically linked sites (Greenblatt 1984; Dooner and Belachew 1989). Within the maize p1 locus, reinsertion occurs at sites ranging from 6-bp to 15-kb from the donor element (Athma et al. 1992; Moreno et al. 1992; Weil et al. 1992). In addition, Ac/Ds elements display strong preferential insertions into gene rich regions and exon/intron sequences (Chen et al. 1987; Bennetzen et al. 1994; Rabinowicz et al. 1999; Conrad and Brutnell 2005; Ahern et al. 2009; Vollbrecht et al. 2010). These features make the Ac/Ds system an effective tool for regional gene tagging in maize and other plants.
Unlike standard transposition, alternative transposition reactions (AT) recruit compatible termini from separate, nearby transposons. A variety of configurations of Ac/Ds termini, such as reverse- and directly oriented ends, were reported to undergo AT. While standard transposition results in a net movement of the transposon, AT can generate larger-scale changes in genome structure. For example, reversed ends transposition (RET) produces deletions, inversions, translocations, and duplications (Zhang and Peterson 2004; Zhang et al. 2006, 2009, 2013; Huang and Dooner 2008). One study identified 3 tandem duplications induced by RET of dhAT-Zm in the maize B73 genome, demonstrating the occurrence of RET during genome evolution (Zhang et al. 2013). The second type of AT, termed sister chromatid transposition (SCT), involves the directly oriented transposon termini located on sister chromatids (Weil and Wessler 1993); SCT can induce the formation of deletions and duplications of varying sizes (Zhang and Peterson 1999). The third configuration, macrotransposon (MTn), is delineated by the external 5′ and 3′ termini of nearby Ac/Ds elements, flanking variable sizes of intertransposon segment (ITS). Excision and reinsertion of an Ac/Ds::MTn was first described by Huang and Dooner (2008); this Ac::MTn included a 6.5-kb ITS containing maize bz1 and stc1 sequences. In contrast to RET or SCT, MTn cannot induce large chromosomal rearrangements or chromosomal breakage (Yu et al. 2010), but only mobilizes the included ITS from one position to another.
In this study, we investigated the transposition activity of 3 Ac::MTn elements in maize. We recovered 10 independent Ac::MTn excision and reinsertion events, and compared features of macrotransposition to that of standard Ac/Ds transposition. In general, Ac::MTn shares similar patterns of transposition activity compared to standard Ac transposition, such as sequence features of excision footprints and formation of 8-bp TSDs flanking the insertion targets. Ac::MTn also prefers insertion into genetically linked and genic sequences. We identified a case of a likely Ac::MTn in the endogenous maize B73 genome, suggesting a historic occurrence of MTn transposition event. We show that Ac/Ds::MTn is capable of mobilizing segments of DNA long enough to include a typical full-length plant gene, and may have contributed to the erosion of gene colinearity in syntenic regions during genome evolution. Our results help better elucidate the various roles of transposable elements in shaping genome evolution.
Materials and methods
Maize stocks and screen
In maize nomenclature, p1 alleles can express color in the pericarp (the maternal tissue surrounding the seed) and/or the cob. The first letter designates pericarp color and the second designates cob color. Thus, the progenitor lines P1-rr::MTn (P1-rr458; P1-rr460; and P1-rr908) have color in both the cob and the pericarp due to the intact p1. The progeny lines p1-ww::MTn have no pigmentation in either cob nor the pericarp due to the loss of p1 function. To screen for the macrotransposition candidates, we crossed the progenitor lines P1-rr::MTn (P1-rr458; P1-rr460; and P1-rr908) with Ac tester lines p1-ww4Co63; rm-3::Ds (Kermicle 1980). The Ac tester line p1-ww4Co63; rm-3::Ds is an r1 mutant line with a Ds insertion. With an active Ac from the P1-ww::MTn, the Ds can be excised from r1 gene to restore the aleurone pigmentation. We screened for seeds or sectors with colorless kernel pericarp indicating loss of p1 function, and then confirmed the Ac activity by screening for purple spots in the aleurone. Seeds or sectors with loss of p1 function but retained Ac activity were considered as candidates of MTn transposition.
Molecular biological methods
Leaf tissue from young plants was collected and ground in liquid nitrogen. Total DNA was prepared by using a modified cetyltrimethylammonium bromide (CTAB) extraction protocol (Porebski et al. 1997). HotMaster Taq polymerase from Eppendorf (Hamburg, Germany) was used in PCR experiments. PCR samples were heated at 94°C for 2 min, followed by 35 cycles of 94°C for 20 s, 60°C for 30 s, and 65°C for 2 min. Another cycle at 65°C is extended for 8 min. Primers used are listed in Supplementary Table 1. For inverse PCR, we first digested genomic DNA with HpyCHIV4, then ligated with T4 DNA ligase (New England Biolabs). First-round PCR with primers 6 and p1_22643f was performed on the ligation mixture, and second-round PCR with primers Ac4508f and p1_22927f on the products of the first PCR. The PCR products were purified by Gel/PCR DNA fragment extraction kit (IBI Scientific, Peosta, IA, USA), and sequenced directly by the DNA Synthesis and Sequencing Facility, Iowa State University using Sanger sequencing. Southern blot was performed according to published protocols (Sambrook et al.1989); the washing stringency used is 0.5% SDS, 0.5×SSC at 60°C. Hybridization probe 15 from the p1 gene has been described in Athma and Peterson (1991).
Genome search for Ac::MTn structures
B73 and NAM lines
We used the 150 bp terminal sequences from Ac 5′ and 3′ termini as query sequences under default setting in a BLAST search against maize B73 reference genome (B73 RefGen_v2) (Schnable et al. 2009). Two separate lists of hits were generated from 2 query termini. The list from Ac 5′ terminus contains 63 hits, and the list from Ac 3′ terminus contains 51 hits. We grouped the hits that are ≤100 kb apart from each other by their map locations among the total of 114 hits. Based on 2 possible termini compositions that are expected in Ac::MTns, we extracted groups that each contain 3–4 hits and include both Ac 5′ and 3′ termini. From the total of 2 hits groups that satisfy the initial search, we observed 1 group including 2 Ac 5′ termini and 1 Ac 3′ terminus in the orientations expected for Ac::MTns. This candidate was further evaluated in the most updated B73 assembly Zm-B73-REFERENCE-NAM-5.0. The sequence from the new genome assembly indicates a double Ds with one element insert into the other in the same orientation. We observed 8-bp TSD sequences flanking the internal Ds, confirming the Ac transposase-induced insertion. We also observed 8-bp TSD flanking the candidate MTn, indicating a historic transposition event of the MTn. We did a BLAST search against the NAM founder lines and identified highly conserved MTn from 5 diverse lines including B97, NC358, NC350, CML 247, and CML322. The TSD sequences are all intact in the NAM lines.
Annotation analysis for p1-ww::MTn alleles
The annotation analyses of the confirmed sequences for 10 p1-ww::MTn alleles include mapping physical and genetic positions and identifying gene models. We blasted against Zm-B73-REFERENCE-NAM-5.0 assembly and IBM2 to obtain the physical positions and genetic coordinates for each Ac::MTn insertion, respectively. We extracted evidence-based gene models for the flanking sequences of each insertion from the official gene models for Zm-B73-REFERENCE-NAM-5.0 (Hufford et al. 2021).
Results
Excision and reinsertion events are identified in p1-ww::MTn alleles
In this study, we used kernel pericarp pigmentation as a phenotypic marker to detect structural changes at the maize p1 locus. The p1 gene encodes an R2R3 Myb transcription factor that regulates flavonoid pigment biosynthesis in pericarp, cob, tassel glumes, and silk (Lechelt et al. 1989; Grotewold et al. 1991, 1994). Pigmentation of pericarp and cob are easily scored, and thus were used to indicate different allele-specific expression patterns of p1: P1-rr specifies red pigmentation in both pericarp and cob, while p1-ww shows no pigmentation in either pericarp or cob (Grotewold et al. 1991, 1994; Athma et al. 1992).
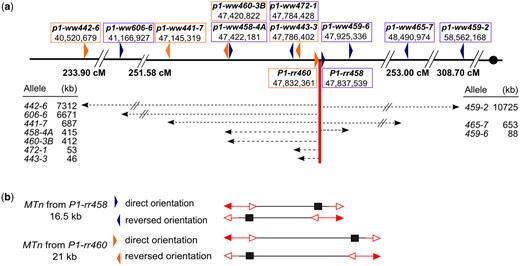
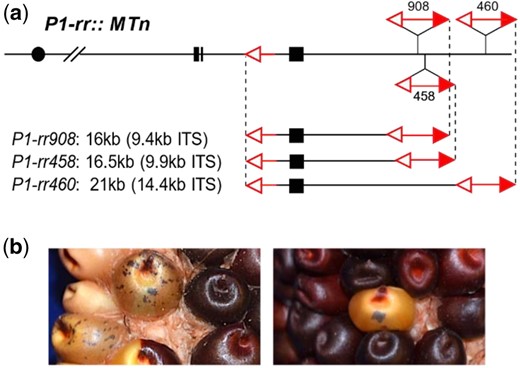
Yu et al. (2010) identified 5 Ac::MTn alleles (P1-ovov455, P1-rr905, P1-rr908, P1-rr458, and P1-rr460) at the maize p1 locus in a previous study. All of the Ac::MTn alleles are derived from the common progenitor p1-vv9D9A and thus their fAc elements are inserted in the same location as that in p1-vv9D9A. Three Ac::MTn alleles were further studied here including P1-rr908, P1-rr458, and P1-rr460. As shown in Fig. 1a, Ac::MTn is composed of the 3′ terminus from fAc at p1 intron 2, and the 5′ terminus from a full-sized Ac element downstream of p1. The internal sequences (ITS) include partial intron 2 and the full sequences of exon 3 of p1. The size of each Ac::MTn is determined by the location of Ac: P1-rr908 is 16 kb in total length, including a 9.4 kb ITS. The Ac in P1-rr458 is 500 bp downstream from Ac in P1-rr908, resulting in a 16.5 kb Ac::MTn with a 9.9 kb ITS. Finally, P1-rr460 contains an Ac inserted farther downstream, resulting in the largest Ac::MTn of 21 kb including a 14.4 kb ITS.
Ac::MTn at p1 locus. a) Schematic structure of Ac::MTn alleles at p1 locus. Filled boxes are maize p1 gene exons 1, 2, and 3 (left to right). open and filled red arrowheads indicate 3′ and 5′ Ac termini, respectively. Three Ac::MTn alleles share a common fAc terminus located in p1 intron 2, and Ac elements (numbered) inserted at 3 different positions downstream of the p1 gene, producing various ITS (inter-transposon segment) sizes. b) Phenotypes of P1-rr::MTn/p1-ww; r1-m3::Ds kernels. Red pigmentation in pericarp and cob is from the expression of p1 gene in P1-rr::MTn allele. Kernels with colorless pericarp arise from loss of p1 function, possibly caused by MTn excision. Purple aleurone sectors resulting from Ds excisions from r1-m3 report active Ac. Kernels with colorless pericarp and purple aleurone sectors were screened as candidates of Ac::MTn excision and reinsertion.
Before transposition, all 3 Ac::MTn alleles retain a functional p1 gene and thus exhibit red pigmentation of both pericarp and cob. However, the excisions of Ac::MTn disrupt p1 by removing exon 3, thus producing new p1-ww excision alleles. To identify cases in which the Ac::MTn had excised and reinserted into a new site, we crossed P1-rr::MTn plants with Ac tester line p1-ww; r1-m3::Ds. If Ac is active in P1-rr::MTn or p1-ww::MTn, then purple sectors on the aleurone can be observed due to reversion of the r1 gene by somatic excision of Ds from the r1-m3 allele (Kermicle 1980). The resulting ears were visually screened to identify candidate Ac::MTn transposition events from single kernel or multikernel sectors with colorless pericarp and purple-spotted kernel aleurone (Fig. 1b). The pedigree of the p1-ww::MTn alleles were summarized in Supplementary Fig. 1.
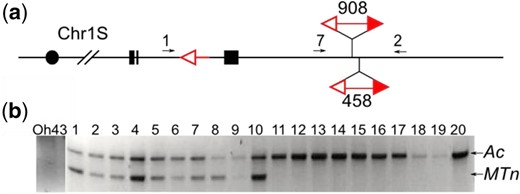
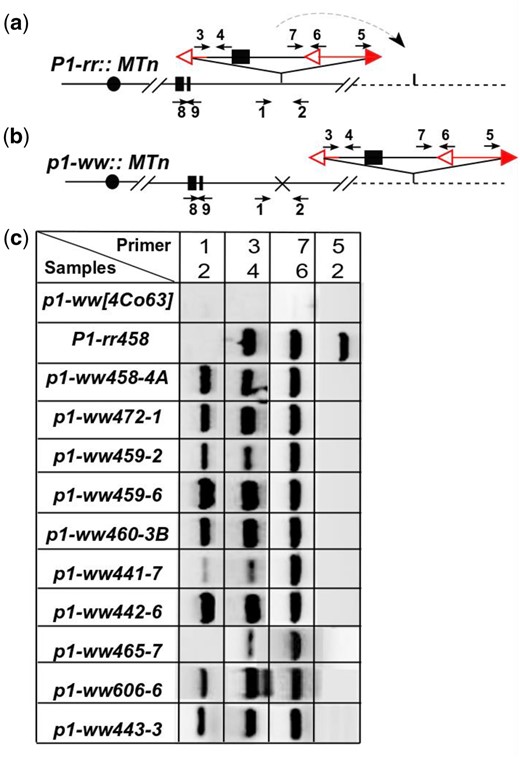
Molecular analysis was then performed on candidates of Ac::MTn transposition obtained from the visual screen (Fig. 2). We first used primers 1 and 2 flanking the Ac::MTn to detect its excision; candidates in which MTn had excised should produce strong bands. To confirm Ac::MTn excision and to exclude somatic excision events, a second PCR amplifying the junction between Ac 5′ terminus and the flanking sequences (primers 2 + 5) was performed. Due to MTn excision, this junction was disrupted, and we expected the absence of any PCR products. The MTn reinsertion events were identified if the ITS sequences were detected by both primers 3 + 4 and primers 6 + 7. Using these diagnostic PCR tests, we identified 21 cases of MTn excision and reinsertion among 177 single or multikernel sectors with colorless pericarp and purple-spotted kernel aleurone from phenotypic screen (Supplementary Table 2). We named those alleles p1-ww::MTn because of the loss of function p1 and the presence of Ac::MTn in the genome. We also identified 27 cases of Ac::MTn excisions without reinsertion, and 129 cases with no Ac::MTn excision. The latter 2 groups are not further studied because of the absence of Ac::MTn in the genome, or the disrupted Ac::MTn due to other mechanisms such as standard Ac insertion or recombination and segregation.
PCR analysis of Ac::MTn transposition events. a and b) Schematic structure of the P1-rr::MTn allele and p1-ww::MTn allele with PCR primers labeled as arrows. Excision footprint was marked by “x” at the Ac::MTn donor site. Vertical lines are insertion target of Ac::MTn. c) Gel analysis of PCRs that identify the excision and reinsertion of Ac::MTn.
Southern blotting was performed to confirm the structures of the p1-ww::MTn alleles (Fig. 3). Genomic DNAs, including the grandparent allele p1-vv9D9A; the parental P1-rr::MTn alleles including P1-rr458, 460, and 908; and the progeny alleles p1-ww::MTn including p1-ww472-1, p1-ww606-6, p1-ww459-6, and p1-ww459-2, were digested with Kpn1 and hybridized with p1 probe 15. The 6.3-kb bands from upstream sequences of p1 are common to all P1-rr::MTn and p1-ww::MTn alleles, indicating that p1 upstream sequences are not involved in the MTn transposition. The 10.6-kb fragments generated from fAc of Ac::MTn and the fAc flanking sequences at p1 locus were observed in all of the P1-rr::MTn alleles, confirming the location of Ac::MTn at p1 locus before transposition. However, this fragment is replaced by a new band of various sizes in p1-ww::MTn alleles including p1-ww472-1, p1-ww606-6, p1-ww459-6, and p1-ww459-2, indicating that the Ac::MTn was excised from p1 and reinserted at new sites at various locations. The sizes of the new bands depend on the location of the new insertion.
Southern blot confirming the location of Ac::MTn reinsertion. a) Schematic structure of the P1-rr::MTn at p1 locus (black line) and p1-ww::MTn at insertion site (dotted line). The expected fragments include a 6.3 kb fragment proximal to p1 and a 10.6 kb fragment from p1 sequences. The 10.6 kb bands are expected to be replaced by a band of new size in p1-ww::MTn lines. b) Result of Southern blot with bands labeled. Lane 2 contains DNA from maize inbred B73, genotype p1-wr, which has a tandem amplification of p1 genes that produces intense band at ∼7 kb (Sekhon et al. 2007), and a single copy band at about 10.6 kb. These bands are also observed in lane 8, which has the allele p1-ww472-1 heterozygous with p1-wr [B73].
To identify the reinsertion sites for the transposed MTn in the confirmed p1-ww::MTn alleles, we performed inverse PCR or Ac casting (Singh et al. 2003; Wang and Peterson 2013) and captured the sequences flanking each reinserted MTn. Putative MTn insertion sites were confirmed by PCR using one primer on the corresponding terminus of MTn and a second primer from the newly captured flanking sequences (Fig. 4). We have recovered reinsertion sites for 10 p1-ww::MTn alleles, all of which were confirmed, including 6 from P1-rr458 and 4 from P1-rr460; none were derived from P1-rr908. With one junction between the MTn terminus (either Ac terminus or fAc terminus) and the flanking sequences confirmed, we extracted sequences from B73 genome assembly, and designed primers to recover the other junction between the new flanking sequences and the remaining MTn terminus. We recovered a total of 8 junctions from the 10 confirmed reinsertions. In 2 cases, the junctions could not be amplified, possibly due to sequence polymorphisms between B73 and the mixed genetic background of the Ac::MTn lines. Among the 8 cases with both flanking junctions, 6 of these contained 8-bp TSDs flanking the transposed Ac::MTn; the 2 remaining cases had 6- and 3-bp TSDs, respectively. The complete flanking sequences of the reinserted MTn are listed in Supplementary Data 1.
PCR verification of Ac::MTn reinsertions. a) Schematic structure of p1-ww::MTn allele with PCR primers labeled. b) Gel analysis of verification PCR. Reinserted locations were confirmed by PCR with primers “Acfl” and “5,” and primers “fAcfl” and “6.” Primers “Acfl” and “fAcfl” are from newly obtained flanking sequences. Lanes “+” use templates from genomic DNA of each allele, and lanes “-” contain DNA from p1-ww [4Co63] as the negative control. Independent Ac::MTn insertions result in different locations in genome, thus primers “Acfl” and “fAcfl” are different in sequences and generate bands with different sizes as observed for each allele.
Ac::MTn preferentially inserts into nearby genic sequences
By aligning the Ac::MTn reinsertion target sequences with the maize B73 reference genome, we found that all insertions are located at the short arm of chromosome 1 (summarized in Fig. 5 and Table 1). The distances of the reinsertion sites from the p1 locus range from 46-kb to 10.7-Mb of physical distance, including 8 distal sites and 2 proximal sites. Moreover, the reinsertions are all genetically linked to p1, including 7 cases that were mapped to the same genetic map position as p1.
Summary of Ac::MTn reinsertions. a) The distribution of Ac::MTn reinsertion sites on chromosome 1S. The original location of Ac::MTn is at 47,832,361 (P1-rr460) or 47,837,539 (P1-rr458). The genetic mapping coordinate numbers labeled along the chromosome are extracted from IBM neighbors v.2 1. Arrows below the chromosome indicate the directions and physical distances of reinserted Ac::MTn in the recovered alleles. b) Orientations of the Ac::MTn reinsertions.
Summary of p1-ww::MTn alleles.
| Allele . | Progenitor . | Excision . | Reinsertion . | ||||
|---|---|---|---|---|---|---|---|
| Footprint . | Insertion site on chr1a . | TSD . | Official gene modelc . | Location in the gene model . | Gene # ± 50 kb flanking . | ||
| p1-ww458-4A | P1-rr458 | Typical | 47422181 | GCGGGGAG | Zm00001eb014140 | Exon | 2 |
| p1-ww472-1 | “ | Typical | 47784428 | NDb | Zm00001eb014210 | 2249 bp distal | 4 |
| p1-ww459-2 | “ | Typical | 58562168 | ND | NA | NA | 1 |
| p1-ww459-6 | “ | Typical | 47925336 | GCTAGC | Zm00001eb014260 | Exon | 5 |
| p1-ww465-7 | “ | Deletion | 48490974 | TGCATGCA | Zm00001eb014490 | 5725 bp distal | 3 |
| p1-ww606-6 | “ | Typical | 41166927 | CAAATTAC | Zm00001eb012530 | Exon | 3 |
| p1-ww460-3B | P1-rr460 | Typical | 47420822 | CTCATCGC | Zm00001eb014140 | Exon | 2 |
| p1-ww441-7 | “ | Typical | 47145319 | CCA | Zm00001eb014060 | Exon | 6 |
| p1-ww442-6 | “ | Typical | 40520679 | CCCAAATC | Zm00001eb012360 | Exon | 2 |
| p1-ww443-3 | “ | Typical | 47786402 | GGCCGGAG | Zm00001eb014210 | 275 bp distal | 4 |
| Allele . | Progenitor . | Excision . | Reinsertion . | ||||
|---|---|---|---|---|---|---|---|
| Footprint . | Insertion site on chr1a . | TSD . | Official gene modelc . | Location in the gene model . | Gene # ± 50 kb flanking . | ||
| p1-ww458-4A | P1-rr458 | Typical | 47422181 | GCGGGGAG | Zm00001eb014140 | Exon | 2 |
| p1-ww472-1 | “ | Typical | 47784428 | NDb | Zm00001eb014210 | 2249 bp distal | 4 |
| p1-ww459-2 | “ | Typical | 58562168 | ND | NA | NA | 1 |
| p1-ww459-6 | “ | Typical | 47925336 | GCTAGC | Zm00001eb014260 | Exon | 5 |
| p1-ww465-7 | “ | Deletion | 48490974 | TGCATGCA | Zm00001eb014490 | 5725 bp distal | 3 |
| p1-ww606-6 | “ | Typical | 41166927 | CAAATTAC | Zm00001eb012530 | Exon | 3 |
| p1-ww460-3B | P1-rr460 | Typical | 47420822 | CTCATCGC | Zm00001eb014140 | Exon | 2 |
| p1-ww441-7 | “ | Typical | 47145319 | CCA | Zm00001eb014060 | Exon | 6 |
| p1-ww442-6 | “ | Typical | 40520679 | CCCAAATC | Zm00001eb012360 | Exon | 2 |
| p1-ww443-3 | “ | Typical | 47786402 | GGCCGGAG | Zm00001eb014210 | 275 bp distal | 4 |
Chromosome location was determined from Zm-B73-REFERENCE-NAM-5.0 assembly.
ND for not determined.
Official gene models from Zm-B73-REFERENCE-NAM-5.0.
Summary of p1-ww::MTn alleles.
| Allele . | Progenitor . | Excision . | Reinsertion . | ||||
|---|---|---|---|---|---|---|---|
| Footprint . | Insertion site on chr1a . | TSD . | Official gene modelc . | Location in the gene model . | Gene # ± 50 kb flanking . | ||
| p1-ww458-4A | P1-rr458 | Typical | 47422181 | GCGGGGAG | Zm00001eb014140 | Exon | 2 |
| p1-ww472-1 | “ | Typical | 47784428 | NDb | Zm00001eb014210 | 2249 bp distal | 4 |
| p1-ww459-2 | “ | Typical | 58562168 | ND | NA | NA | 1 |
| p1-ww459-6 | “ | Typical | 47925336 | GCTAGC | Zm00001eb014260 | Exon | 5 |
| p1-ww465-7 | “ | Deletion | 48490974 | TGCATGCA | Zm00001eb014490 | 5725 bp distal | 3 |
| p1-ww606-6 | “ | Typical | 41166927 | CAAATTAC | Zm00001eb012530 | Exon | 3 |
| p1-ww460-3B | P1-rr460 | Typical | 47420822 | CTCATCGC | Zm00001eb014140 | Exon | 2 |
| p1-ww441-7 | “ | Typical | 47145319 | CCA | Zm00001eb014060 | Exon | 6 |
| p1-ww442-6 | “ | Typical | 40520679 | CCCAAATC | Zm00001eb012360 | Exon | 2 |
| p1-ww443-3 | “ | Typical | 47786402 | GGCCGGAG | Zm00001eb014210 | 275 bp distal | 4 |
| Allele . | Progenitor . | Excision . | Reinsertion . | ||||
|---|---|---|---|---|---|---|---|
| Footprint . | Insertion site on chr1a . | TSD . | Official gene modelc . | Location in the gene model . | Gene # ± 50 kb flanking . | ||
| p1-ww458-4A | P1-rr458 | Typical | 47422181 | GCGGGGAG | Zm00001eb014140 | Exon | 2 |
| p1-ww472-1 | “ | Typical | 47784428 | NDb | Zm00001eb014210 | 2249 bp distal | 4 |
| p1-ww459-2 | “ | Typical | 58562168 | ND | NA | NA | 1 |
| p1-ww459-6 | “ | Typical | 47925336 | GCTAGC | Zm00001eb014260 | Exon | 5 |
| p1-ww465-7 | “ | Deletion | 48490974 | TGCATGCA | Zm00001eb014490 | 5725 bp distal | 3 |
| p1-ww606-6 | “ | Typical | 41166927 | CAAATTAC | Zm00001eb012530 | Exon | 3 |
| p1-ww460-3B | P1-rr460 | Typical | 47420822 | CTCATCGC | Zm00001eb014140 | Exon | 2 |
| p1-ww441-7 | “ | Typical | 47145319 | CCA | Zm00001eb014060 | Exon | 6 |
| p1-ww442-6 | “ | Typical | 40520679 | CCCAAATC | Zm00001eb012360 | Exon | 2 |
| p1-ww443-3 | “ | Typical | 47786402 | GGCCGGAG | Zm00001eb014210 | 275 bp distal | 4 |
Chromosome location was determined from Zm-B73-REFERENCE-NAM-5.0 assembly.
ND for not determined.
Official gene models from Zm-B73-REFERENCE-NAM-5.0.
To further understand the insertion preferences relative to the gene space, we extracted evidence-based gene models at the positions of reinsertions from the annotated B73 genome Zm-B73-REFERENCE-NAM-5.0 (Hufford et al. 2021). Nine out of 10 Ac::MTn insertions are located in or near genes, including 6 cases inserted within exon sequences, and 3 cases inserted distal to an evidence-based gene model. For example, the insertion of p1-ww441-7 is located in the exon sequences of gene model Zm00001eb014060, which is 37.45 kb distal to Zm00001eb014070; this site is also 7.3, 11.20, 34.86, and 34.93 kb proximal to Zm00001eb014050, Zm00001eb014040, Zm00001eb014030 and Zm00001eb014020, respectively. Similarly, the Ac::MTn insertion of p1-ww459-6 is located in the exon sequences of the evidence-based gene model Zm00001eb014260; 4 other gene models are located within the flanking ±50 kb region. The MTn of allele p1-ww443-3 is not directly inserted into a gene, but is located 275 bp distal of the gene model Zm00001eb014210. The insertion site is 5.12, 44.55, 45.93, and 50.49 kb distal to Zm00001eb014220, Zm00001eb014230, Zm00001eb014240, and Zm00001eb014250, respectively. In general, the Ac::MTn insertions show preference for genetically linked sequences. Most of the Ac::MTn insertions occurred into, or near, gene sequences. This insertion preference in the genic sequences is similar to that reported for standard Ac/Ds transposition (Chen et al. 1987; Bennetzen et al. 1994; Rabinowicz et al. 1999; Conrad and Brutnell 2005; Ahern et al. 2009; Vollbrecht et al. 2010). This conclusion is tempered somewhat by the possible differences between B73 and the mixed genetic background of the Ac::MTn lines; however, we did not observe any indication of differing insertion site preferences between Ac/Ds standard transposition and Ac::MTn insertion.
Somatic transposition of Ac::MTn is not detected in P1-rr908
To study the frequency of Ac::MTn transposition, we compared alleles P1-rr908 and P1-rr458. The 2 Ac::MTns differ by only 500 bp, thus enabling use of the same PCR primers (primers 1 + 2, primers 7 + 2 in Fig. 6a) to detect excisions of the Ac::MTn and the component Ac for both alleles. Both P1-rr908 and P1-rr458 alleles were made heterozygous with inbred line Oh43, which does not produce a background band from either primer pair. Shown in Fig. 6b, bands from standard Ac transpositions were observed from both P1-rr908 and P1-rr458 alleles, indicating that the component Ac is somatically active in both lines. However, bands from Ac::MTn excision were absent in all of the sibling plants of P1-rr908 and were only observed in plants of P1-rr458. The result also agrees with the zero recovery of Ac::MTn transpositions from P1-rr908 described previously, indicating that Ac::MTn excision in P1-rr908 is severely repressed.
Somatic transposition of Ac and Ac::MTn in P1-rr458 and P1-rr908 alleles. a) Schematic structure of the P1-rr458 and P1-rr908 alleles. Numbered arrows indicate primers designed to detect somatic transposition of Ac and Ac::MTn. b) PCR Gel analysis. DNA templates in lanes 1–10 are from sibling plants of P1-rr458/Oh43; Lanes 11–20 are from sibling plants of P1-rr908/Oh43. Lane “Oh43” contains genomic DNA from Oh43 as template serving as a negative control. The top bands are produced by primers “7” and “2,” from Ac transposition events; while the bottom bands are from primers “1” and “2,” from Ac::MTn excisions.
Identification of historic transposed MTn in B73 genome and NAM lines
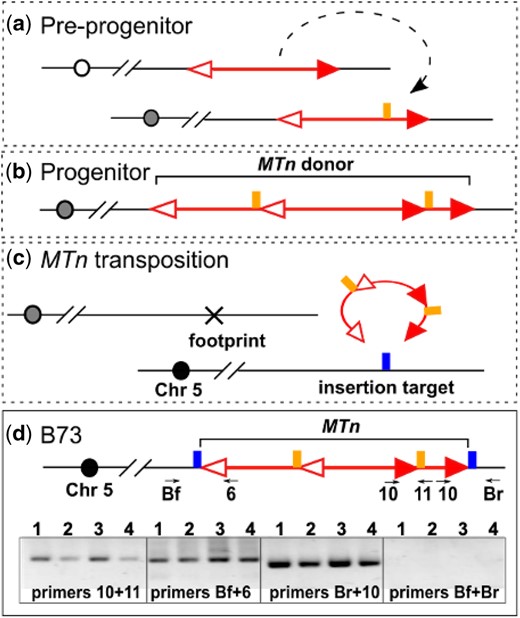
We also investigated the maize B73 reference genome for evidence of possible MTn transposition events occurring during maize genome evolution. One such case is from chromosome 5 as shown in Fig. 7; corresponding sequences are presented in Supplementary Data 2. The potential MTn identified here consists of a 4564-bp Ds inserted in another 4,564 bp Ds at a location 637-bp from the 5′ end of the hosting Ds in the same orientation. Both Ds elements share 99% sequence similarity with Ac sequence. Using PCR of genomic maize B73 DNA, we isolated the internal junctions between 2 Ds elements, as well as the junctions between MTn and flanking sequences, confirming the structure of the B73 MTn. By comparing the sequences flanking the internal Ds that is inserted in the host Ds, we observed 8-bp TSD flanking the insertion Ds (AGTGGAGG), indicating that this MTn was formed through Ac/Ds transposition. By comparing the MTn flanking sequences, we observed 8-bp TSD sequences (CCTACGAC) adjacent to the 5′ and 3′ temini of MTn, strongly indicating that this MTn was transposed to the current location in B73 from its original location in an ancient line. The ancient transposition would also leave an excision footprint sequence at the MTn donor site; however, this origin locus may have been segregated from the insertion site in the ancient line and would be difficult to trace.
MTn in B73 genome. a) Predicted preprogenitor allele with 2 Ds elements inserted into one another at the target site (vertical line). b) The structure of MTn in the progenitor line of B73. TSDs (2 vertical lines) flank the internal Ds. c) The ancient transposition of MTn from the donor site to the target site (vertical line). d) MTn in B73. Upper panel: MTn is located on chromosome 5 of B73 genome, flanked by TSDs (2 vertical line). Numbered arrows indicate primers used in PCR to detect somatic transposition of MTn. Lower panel: PCR results testing the structure of MTn and its activity. Lane1-4: B73 carrying active Ac from p1-vv9D9A; B73 carrying active Ac from p1-wwdef1; B73; B73 (the latter 2 samples are from different DNA preparations).
However, somatic transposition of the existing B73 MTn was undetectable, indicating that this MTn is no longer active to transpose in B73. This was tested by crossing B73 with the active Ac line p1-vv9D9A (Athma et al. 1992) or p1-ww-def1 (Zhang and Peterson 1999), then performing PCR using primers flanking the MTn. No PCR products from MTn excision were observed in the progeny of either cross, indicating that somatic transposition of the B73 MTn is in some way repressed, even in the presence of an active Ac in trans. A search on the data from NAM Consortium (Hufford, et al., 2021) indicates that B73 MTn is heavily methylated at CG and CHG (Supplementary Fig. 2). A search on the data of B73 small RNAs (Zuo et al. 2016; Wang et al. 2020) also identified small RNAs that are homologous to the terminal sequences of the Ds elements in MTn (Supplementary Fig. 3). Therefore the MTn probably lost transposition activity due to epigenetic silencing mechanism (Leu et al. 1992; Brutnell and Dellaporta 1994).
We have also identified a highly conserved MTn in 5 NAM founder lines including B97, NC358, NC350, CML 247 and CML322. The sequences in MTn in NAM lines are >99.90% identical to the element observed in B73, all including the intact TSD sequences flanking the insertion Ds and TSD sequences flanking MTn (Supplementary Data 3 and 4). The occurrence of MTn elements in diverse lines suggests the ancient formation and transposition of MTn.
Discussion
Ac::MTn transposition shares similar features with Ac/Ds standard transposition
In this study, we used the p1 gene as a visual marker to screen pigmented maize stocks containing P1-rr::MTn alleles for loss-of-function alleles (p1-ww) carrying transposed Ac::MTn. The P1-rr::MTn alleles contain fAc inserted in p1 and Ac inserted downstream of p1; transpositions of Ac::MTn separate p1 exon 3 from the p1 promoter and exons 1 and 2. To diagnose the transposition, we used 4 PCR assays to detect the presence or absence of external and internal junctions, and thereby identified 10 Ac::MTn transposition events. By using inverse PCR or Ac casting, we further cloned the reinsertion site for each event. Reinsertions of Ac::MTn were plotted and also annotated on the maize B73 genome assembly.
Two typical cases of Ac::MTn transposition are illustrated in Supplementary Fig. 4. Due to the catalysis by Ac TPase, the transposition of Ac::MTn shares similar features with standard Ac transposition:
(1) Ac::MTn produces a similar pattern of excision footprint at Ac::MTn donor site. By sequencing the excision band in PCR using primers 1 and 2 in Fig. 2c, we observed minor changes on nucleotides flanking the Ac::MTn donor site in 9 out of 10 excision cases, which is typical in an Ac TPase-induced transposition (Supplementary Fig. 5). The p1-ww465-7 allele is an exception in that it shows no band from the footprint PCR assay. The p1 sequences outside of Ac::MTn are also absent (Supplementary Fig. 6); these observations may due to a possible deletion at p1.
(2) Upon re-insertion, Ac::MTn generates 8-bp TSDs flanking the transposed Ac::MTn in 6 of 8 cases; a 6-bp TSD in one case, and a 3-bp TSD in one case. We compared the frequencies of perfect and imperfect TSDs generated by Ac::MTn insertions with that of 109 Ds transpositions from the a1 locus of maize (Vollbrecht et al. 2010), and observed no significant differences between Ac::MTn and Ds transpositions; Fisher-Exact and Barnard’s Test (P > 0.05) (Supplementary Data 5).
(3) Similar to Ac/Ds transposition (Athma et al. 1992; Moreno et al. 1992; Weil et al. 1992; Vollbrecht et al. 2010), Ac::MTn preferentially inserts into linked sites in or near genes. Nine of 10 Ac::MTn reinsertions occur at sites genetically linked to the Ac::MTn donor site in the p1 locus, and 6 of 10 reinsertions are in the exons of a gene model.
Conclusively, the transposition of Ac::MTn shows a similar pattern with standard Ac/Ds transposition in regard of the patterns of excision footprint sequences, TSD sequences, and the preferential insertion targets.
The mechanism that silences Ac::MTn in P1-rr908 is unclear
It is interesting that the smallest Ac::MTn allele P1-rr908 (16-kb) shows no detectable somatic MTn excision, even though its component Ac shows an apparently normal frequency of somatic transposition. In an attempt to understand the repression or silencing of Ac::MTn in P1-rr908, we first sequenced the Ac::MTn termini and found them to be intact and unchanged from the component Ac and fAc (Supplementary Data 6). Thus we eliminated the possibilities of Ac::MTn junction integrity for the silencing of MTn in P1-rr908.
We then compared the GC content of the 500 bp sequences flanking the Ac of the Macrotransposons (MTns) P1-rr908, P1-rr458, and P1-rr460 because GC content correlates with the possible CpG islands that can be the target of methylation. Both P1-rr908 and P1-rr458 contain a high GC% (GC% = 75%) in the sequences flanking MTn 5′ terminus (Supplementary Fig. 7, Supplementary Data 7), compared to the 46.5% overall GC content in maize genes (Haberer et al. 2005), making them possible to establish CpG islands. Due to the comparable GC contents flanking Ac::MTn in both P1-rr908 and P1-rr458, we cannot explain the varied MTn activity between P1-rr908 and P1-rr458. Further tests on the methylation status at MTn locus of each line will be required to clarify this question.
Previous studies consistently showed an inverse relationship between the frequencies of AT-induced chromosome breakage/rearrangements and the distances separating the participating transposon termini (Huang and Dooner 2008; Yu et al. 2010). Due to the inactive Ac::MTn in P1-rr908, we are unable to compare it with other active Ac::MTn to study the effect of MTn sizes on their activities. However, our results do indicate that, in addition to the effects of MTn size on transposition frequency, there are other factor(s) such as epigenetic modifications that may exert a dominant repressive effect on MTn transposition activity.
Occurrence of MTn transposition events during maize genome evolution
Using bioinformatics searches we identified an MTn-like structure flanked by 8-bp TSDs in the maize lines B73, B97, NC358, NC 350, CML 247, and CML 322. The MTn was derived from a progenitor locus containing one Ds inserted into another Ds in the same orientation. Another instance of Ac/Ds self-insertion comprises the chromosome-breaking doubleDs, which consists of one Ds element inserted into a second Ds in the reversed orientation (McClintock 1948; Döring and Starlinger 1984). Similar structures have been characterized including Sesqui-Ac (Martínez-Férez and Dooner 1997) and half-double Ds (English et al. 1995), both of which show chromosome breaking activity similar to that of doubleDs. However, the chromosome breaking self-insertions that were described above have their transposition-competent termini in opposite orientations. In this configuration, chromosome breakage is thought to arise from sister chromatid fusions induced by transposition reactions that target termini located on sister chromatids following DNA replication (Weil and Wessler 1993; Yu et al. 2010). In the MTns studied here, the involved Ds sequences are in the same orientation and thus transposition can occur at the 3′ fAc and 5′ Ds termini, resulting in mobilization of the entire compound structure. Tests have shown that these MTns cannot induce chromosomal breakage (Yu et al. 2010), and thus may be more stable over evolutionary time.
The identification of the MTn in diverse maize lines demonstrates that MTn transposition occurred very early during maize genome evolution. However, this MTn appears to be currently incapable of transposition, even in the presence of a known active Ac element introduced in trans. The Ds contained in the MTn share 99% identity to the corresponding regions in a canonical Ac, suggesting that the silencing is not likely due to sequence polymorphisms, but rather to epigenetic effects. For example, DNA methylation is correlated with silencing of Ac elements at wx-m7 and p1 loci (Chomet et al. 1987; Brutnell and Dellaporta 1994; Conrad and Brutnell 2005).
Ac::MTn can serve as a potential genome-engineering tool
Some features of Ac::MTn transposition may be applied for purposes of genome engineering. First, the Ac::MTn shows a high capacity to carry plant genes: various studies have demonstrated the ability of Ac::MTn to transpose ITSs of 6.5-kb (Huang and Dooner 2008), 9.9 and 14.4-kb (this study), which is similar to the size range of typical plant genes. The tendency of Ac::MTn to undergo local transposition into nearby genic sequences demonstrated in this study suggests that engineered MTns could be useful tools for locus interaction and epigenetics studies. For example, Ac::MTn constructs could carry full-size genes, large and complex regulatory elements, and hypo- or hyper-methylated sequence islands. Subsequent independent Ac::MTn transpositions could mobilize the foreign gene sequences into a variety of positions in or near a nearby gene, in both orientations. Alterations in expression of the target gene can reveal parameters of regulatory interactions such as the importance of distance and orientation, as well as epigenetic effects. A recent study showed that multiple recurrent inversions generated by independent Ac alternative transposition events induced varying levels of ectopic expression of a normally silent nearby gene (Sharma et al. 2021). Furthermore, it would be straightforward to modify the internal Ac termini to create “stabilized Ac” forms (Hehl and Baker 1989), that would eliminate single Ac transposition but retain full Ac::MTn transposition capability.
MTn may contribute to the formation of dispersed transduplicated genes
Comparative studies of Arabidopsis and its relatives have shown that a large proportion (from one-fourth to three-fourths) of genes in Arabidopsis have been transposed during evolution in the Brassicales and Rosids (Freeling et al. 2008; Woodhouse et al. 2011). Maize is also known to contain a large proportion of transduplicated genes, some of which are carried by MULE and helitrons transposons (Jiang et al. 2004; Juretic et al. 2005; Lisch 2005; Morgante et al. 2005; ). Here, we show that an Ac::MTn can acquire and mobilize large segments of DNA capable of containing entire genes, including introns and flanking regulatory sequences. These results suggest that MTn transposition may have contributed to gene duplication and dispersal during the evolution of maize and other plants.
Data availability
The Ac::MTn insertion data analyzed here are under NCBI accession numbers OM650318, OM650319, OM650320, OM650321, OM650322, OM650323, OM650324, OM650325, OM650326, OM650327, OM650328, OM650329, OM650330, OM650331, OM650332, OM650333, OM650334, and OM650335. Maize stocks are available upon request. All data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material is available at GENETICS online.
Acknowledgments
The authors thank Tao Zuo for extracting Ac termini sequences from maize B73 genome assembly. We thank Terry Olson, Alina Ott, and Pooja Gupta for technical assistance. They also thank Douglas Baker for field assistance.
DW, CY, JZ, and TP conceived and designed the experiments; DW performed the experiments; DW and TP wrote the paper.
Funding
This study is supported by the USDA National Institute of Food and Agriculture Hatch project number IOW05282 and IOW05669, by State of Iowa funds, and by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under Grant #P20GM103476.
Conflicts of interest
The authors declare no conflict of interest.



![Southern blot confirming the location of Ac::MTn reinsertion. a) Schematic structure of the P1-rr::MTn at p1 locus (black line) and p1-ww::MTn at insertion site (dotted line). The expected fragments include a 6.3 kb fragment proximal to p1 and a 10.6 kb fragment from p1 sequences. The 10.6 kb bands are expected to be replaced by a band of new size in p1-ww::MTn lines. b) Result of Southern blot with bands labeled. Lane 2 contains DNA from maize inbred B73, genotype p1-wr, which has a tandem amplification of p1 genes that produces intense band at ∼7 kb (Sekhon et al. 2007), and a single copy band at about 10.6 kb. These bands are also observed in lane 8, which has the allele p1-ww472-1 heterozygous with p1-wr [B73].](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/genetics/221/4/10.1093_genetics_iyac067/1/m_iyac067f3.jpeg?Expires=1716381013&Signature=3Wy239toMiWgfdDifBMDiCEs6VjBsJ7A5pa981WwlJhQvuJ5WRAe7ZTwjUKGx8RP05VwVcePI3qPxMLdLTXlmOI5o6VSxvne2H~tcpsz2GO6MVTkahM9mAjh45wrARtZ~3cgW7EbXn8yNEJFB0O7PUOlgXjeh1zATTj6HvL4uWERW97wodYzHLGwvHGTDyEd8VCPxnkjocyNcwVfFM4ubaxT6SCbD2YEG-7gmiSaEEFuxSttg~adFe5VsNUts36NeKUL74GCeM8-0BfQFpffSX2Ree5eSkkxPSIS-8UZFRjWsVu6EM6WGlww40vhv5sw2TtPuyvsT940ryCXsis7qw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![PCR verification of Ac::MTn reinsertions. a) Schematic structure of p1-ww::MTn allele with PCR primers labeled. b) Gel analysis of verification PCR. Reinserted locations were confirmed by PCR with primers “Acfl” and “5,” and primers “fAcfl” and “6.” Primers “Acfl” and “fAcfl” are from newly obtained flanking sequences. Lanes “+” use templates from genomic DNA of each allele, and lanes “-” contain DNA from p1-ww [4Co63] as the negative control. Independent Ac::MTn insertions result in different locations in genome, thus primers “Acfl” and “fAcfl” are different in sequences and generate bands with different sizes as observed for each allele.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/genetics/221/4/10.1093_genetics_iyac067/1/m_iyac067f4.jpeg?Expires=1716381013&Signature=mJj4hzM9nGQyyLa~MjTk3h9nhjDUc3mAAtDjgKRsyIF3kVysU8mLY3~1AP1dqZouaA9NBB~aTuGg3ZcdnJML7gC0uYDN710akCIWoSY3-eaqrpRcpYMd6a3HEDUGMnpd4y4~K9q9uxTa1uOmN8iZpoA7PYxrap2iSx4kHyWSh9fHTEs5v4cEQPsr8MigAyc8pczhv2uF301sKeWeWLm56EkP-S45-8snqYn9p-mCG3lrcbrz9-JFSv4oBpOxjkpyu~7nbsYyzjFMXEZ5W6FIHYzP1nFv-ZTrABQsr9xUEv~12M2FdWccFTtf6UZtk4a1WJ~FSlxnO~l2txSbwR0gmQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)